6KHD
 
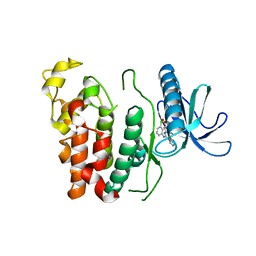 | | Crystal structure of CLK1 in complex with CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK1 | | Authors: | Lee, J.Y, Yun, J.S, Jin, H, Chang, J.H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Selective Inhibition of Cdc2-Like Kinases by CX-4945.
Biomed Res Int, 2019, 2019
|
|
8K06
 
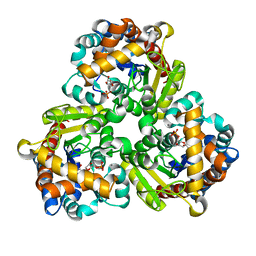 | | Pseudouridine 5'-monophosphate glycosylase from Arabidopsis thaliana -- PSU, R5P bound K185A mutant | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, MANGANESE (II) ION, PSEUDOURIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Lee, J.Y, Kim, S.H, Rhee, S.K. | | Deposit date: | 2023-07-07 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Structure and function of the pseudouridine 5'-monophosphate glycosylase PUMY from Arabidopsis thaliana.
Rna Biol., 21, 2024
|
|
8K05
 
 | |
8K07
 
 | |
5GPC
 
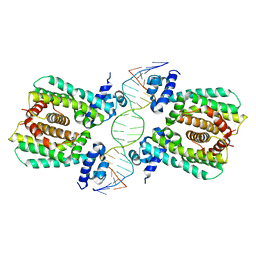 | | Structural analysis of fatty acid degradation regulator FadR from Bacillus halodurans | | Descriptor: | DNA (5'-D(P*CP*AP*TP*GP*AP*AP*TP*GP*AP*GP*TP*AP*TP*TP*CP*AP*TP*TP*CP*AP*T)-3'), DNA (5'-D(P*GP*AP*TP*GP*AP*AP*TP*GP*AP*AP*TP*AP*CP*TP*CP*AP*TP*TP*CP*AP*T)-3'), Transcriptional regulator (TetR/AcrR family) | | Authors: | Lee, J.Y, Yeo, H.K, Park, T.W. | | Deposit date: | 2016-08-01 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of operator sites recognition and effector binding in the TetR family transcription regulator FadR.
Nucleic Acids Res., 45, 2017
|
|
5GP9
 
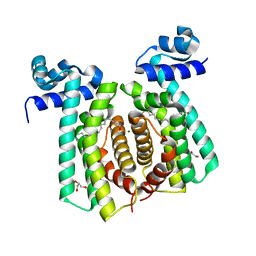 | | Structural analysis of fatty acid degradation regulator FadR from Bacillus halodurans | | Descriptor: | GLYCEROL, MAGNESIUM ION, PALMITIC ACID, ... | | Authors: | Lee, J.Y, Yeo, H.K, Park, Y.W. | | Deposit date: | 2016-08-01 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Structural basis of operator sites recognition and effector binding in the TetR family transcription regulator FadR.
Nucleic Acids Res., 45, 2017
|
|
5GPA
 
 | |
4MF9
 
 | |
4MGF
 
 | |
3KT7
 
 | | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, GLYCEROL, ... | | Authors: | Kim, H.S, Kim, H.L, Kim, K.H, Kim, D.J, Lee, S.J, Yoon, J.Y, Yoon, H.J, Lee, H.Y, Park, S.B, Kim, S.-J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2009-11-24 | | Release date: | 2010-01-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex
Nucleic Acids Res., 38, 2010
|
|
3KT4
 
 | | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex | | Descriptor: | FE (III) ION, PKHD-type hydroxylase TPA1 | | Authors: | Kim, H.S, Kim, H.L, Kim, K.H, Kim, D.J, Lee, S.J, Yoon, J.Y, Yoon, H.J, Lee, H.Y, Park, S.B, Kim, S.-J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2009-11-24 | | Release date: | 2010-01-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex
Nucleic Acids Res., 38, 2010
|
|
3KT1
 
 | | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex | | Descriptor: | FE (III) ION, GLYCEROL, PKHD-type hydroxylase TPA1, ... | | Authors: | Kim, H.S, Kim, H.L, Kim, K.H, Kim, D.J, Lee, S.J, Yoon, J.Y, Yoon, H.J, Lee, H.Y, Park, S.B, Kim, S.-J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2009-11-24 | | Release date: | 2010-01-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex
Nucleic Acids Res., 38, 2010
|
|
3RYO
 
 | | Crystal Structure of Enhanced Intracellular Survival (Eis) Protein from Mycobacterium tuberculosis with Acetyl CoA | | Descriptor: | ACETYL COENZYME *A, Enhanced intracellular survival protein | | Authors: | Kim, K.H, An, D.R, Yoon, J.Y, Kim, H.S, Yoon, H.J, Song, J, Im, H.N, Kim, J, Kim, D.J, Lee, S.J, Kim, H.J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2011-05-11 | | Release date: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7
Proc.Natl.Acad.Sci.USA, 2012
|
|
7RSJ
 
 | | Structure of the VPS34 kinase domain with compound 14 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-{4-[(7R,8R)-4-oxo-7-(propan-2-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazin-2-yl]pyridin-2-yl}cyclopropanecarboxamide, ... | | Authors: | Hu, D.X, Patel, S, Chen, H, Wang, S, Staben, S, Dimitrova, Y.N, Wallweber, H.A, Lee, J.Y, Chan, G.K.Y, Sneeringer, C.J, Prangley, M.S, Moffat, J.G, Wu, C, Schutt, L.K, Salphati, L, Pang, J, McNamara, E, Huang, H, Chen, Y, Wang, Y, Zhao, W, Lim, J, Murthy, A, Siu, M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | Structure-Based Design of Potent, Selective, and Orally Bioavailable VPS34 Kinase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7RSP
 
 | | Structure of the VPS34 kinase domain with compound 14 | | Descriptor: | (7R,8R)-2-[(3R)-3-methylmorpholin-4-yl]-7-(propan-2-yl)-6,7-dihydropyrazolo[1,5-a]pyrazin-4(5H)-one, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3 | | Authors: | Hu, D.X, Patel, S, Chen, H, Wang, S, Staben, S, Dimitrova, Y.N, Wallweber, H.A, Lee, J.Y, Chan, G.K.Y, Sneeringer, C.J, Prangley, M.S, Moffat, J.G, Wu, C, Schutt, L.K, Salphati, L, Pang, J, McNamara, E, Huang, H, Chen, Y, Wang, Y, Zhao, W, Lim, J, Murthy, A, Siu, M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure-Based Design of Potent, Selective, and Orally Bioavailable VPS34 Kinase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7RSV
 
 | | Structure of the VPS34 kinase domain with compound 5 | | Descriptor: | (5aS,8aR,9S)-2-[(3R)-3-methylmorpholin-4-yl]-5,5a,6,7,8,8a-hexahydro-4H-cyclopenta[e]pyrazolo[1,5-a]pyrazin-4-one, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3, ... | | Authors: | Hu, D.X, Patel, S, Chen, H, Wang, S, Staben, S, Dimitrova, Y.N, Wallweber, H.A, Lee, J.Y, Chan, G.K.Y, Sneeringer, C.J, Prangley, M.S, Moffat, J.G, Wu, C, Schutt, L.K, Salphati, L, Pang, J, McNamara, E, Huang, H, Chen, Y, Wang, Y, Zhao, W, Lim, J, Murthy, A, Siu, M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-Based Design of Potent, Selective, and Orally Bioavailable VPS34 Kinase Inhibitors.
J.Med.Chem., 65, 2022
|
|
2WH9
 
 | | Solution structure of GxTX-1E | | Descriptor: | GUANGXITOXIN-1EGXTX-1E | | Authors: | Lee, S.K, Jung, H.H, Lee, J.Y, Lee, C.W, Kim, J.I. | | Deposit date: | 2009-05-02 | | Release date: | 2010-05-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Gxtx-1E, a High Affinity Tarantula Toxin Interacting with Voltage Sensors in Kv2.1 Potassium Channels.
Biochemistry, 49, 2010
|
|
1IX1
 
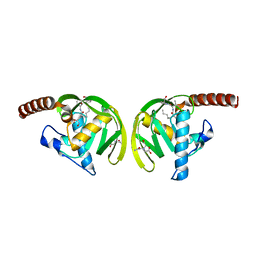 | | Crystal Structure of P.aeruginosa Peptide deformylase Complexed with Antibiotic Actinonin | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, ACTINONIN, ZINC ION, ... | | Authors: | Kim, H.-W, Yoon, H.-J, Lee, J.Y, Han, B.W, Yang, J.K, Lee, B.I, Ahn, H.J, Lee, H.H, Suh, S.W. | | Deposit date: | 2002-06-07 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of peptide deformylase from Staphylococcus aureus in complex with actinonin, a naturally occurring antibacterial agent
Proteins, 57, 2004
|
|
3SXN
 
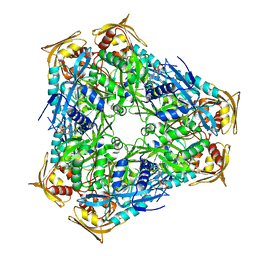 | | Mycobacterium tuberculosis Eis protein initiates modulation of host immune responses by acetylation of DUSP16/MKP-7 | | Descriptor: | COENZYME A, Enhanced intracellular survival protein | | Authors: | Kim, K.H, An, D.R, Yoon, J.Y, Kim, H.S, Yoon, H.J, Song, J.S, Im, H.N, Kim, J, Kim, D.J, Lee, S.J, Kim, H.J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2011-07-15 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GSQ
 
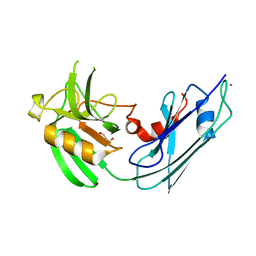 | | Structural basis for the inhibition of Mycobacterium tuberculosis L,D-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains | | Descriptor: | CALCIUM ION, GLYCEROL, Probable conserved lipoprotein LPPS | | Authors: | Kim, H.S, Kim, J, Im, H.N, Yoon, J.Y, An, D.R, Yoon, H.J, Kim, J.Y, Min, H.K, Kim, S.-J, Lee, J.Y, Han, B.W, Suh, S.W. | | Deposit date: | 2012-08-28 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the inhibition of Mycobacterium tuberculosis L,D-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GSR
 
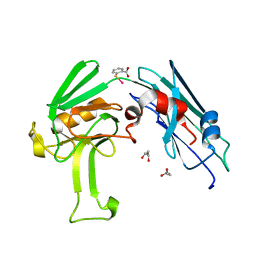 | | Structural basis for the inhibition of Mycobacterium tuberculosis L,D-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains | | Descriptor: | 2-(ETHYLMERCURI-THIO)-BENZOIC ACID, GLYCEROL, Probable conserved lipoprotein LPPS | | Authors: | Kim, H.S, Kim, J, Im, H.N, Yoon, J.Y, An, D.R, Yoon, H.J, Kim, J.Y, Min, H.K, Kim, S.-J, Lee, J.Y, Han, B.W, Suh, S.W. | | Deposit date: | 2012-08-28 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural basis for the inhibition of Mycobacterium tuberculosis L,D-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GSU
 
 | | Structural basis for the inhibition of Mycobacterium tuberculosis L,D-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains | | Descriptor: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Probable conserved lipoprotein LPPS | | Authors: | Kim, H.S, Kim, J, Im, H.N, Yoon, J.Y, An, D.R, Yoon, H.J, Kim, J.Y, Min, H.K, Kim, S.-J, Lee, J.Y, Han, B.W, Suh, S.W. | | Deposit date: | 2012-08-28 | | Release date: | 2013-02-27 | | Last modified: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the inhibition of Mycobacterium tuberculosis L,D-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3SXO
 
 | | Mycobacterium tuberculosis Eis protein initiates modulation of host immune responses by acetylation of DUSP16/MKP-7 | | Descriptor: | Enhanced intracellular survival protein | | Authors: | Kim, K.H, An, D.R, Yoon, J.Y, Kim, H.S, Yoon, H.J, Song, J.S, Im, H.N, Kim, J, Kim, D.J, Lee, S.J, Kim, H.J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2011-07-15 | | Release date: | 2012-07-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mycobacterium tuberculosis Eis protein initiates modulation of host immune responses by acetylation of DUSP16/MKP-7
To be Published
|
|
1J9L
 
 | | CRYSTAL STRUCTURE OF SURE PROTEIN FROM T.MARITIMA IN COMPLEX WITH VANADATE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, STATIONARY PHASE SURVIVAL PROTEIN, ... | | Authors: | Suh, S.W, Lee, J.Y, Kwak, J.E, Moon, J. | | Deposit date: | 2001-05-28 | | Release date: | 2001-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and functional analysis of the SurE protein identify a novel phosphatase family.
Nat.Struct.Biol., 8, 2001
|
|
1J9J
 
 | | CRYSTAL STRUCTURE ANALYSIS OF SURE PROTEIN FROM T.MARITIMA | | Descriptor: | MAGNESIUM ION, STATIONARY PHASE SURVIVAL PROTEIN, SULFATE ION | | Authors: | Suh, S.W, Lee, J.Y, Kwak, J.E, Moon, J. | | Deposit date: | 2001-05-27 | | Release date: | 2001-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and functional analysis of the SurE protein identify a novel phosphatase family.
Nat.Struct.Biol., 8, 2001
|
|
