4EDA
 
 | | Structures of monomeric hemagglutinin and its complex with an Fab fragment of a neutralizing antibody that binds to H1 subtype influenza viruses: molecular basis of infectivity of 2009 pandemic H1N1 influenza A viruses | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Park, Y.H, Khan, T.G, Lee, J.Y, Kang, S.H, Alam, I. | | Deposit date: | 2012-03-27 | | Release date: | 2013-05-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Insight into structural diversity of influenza virus haemagglutinin
J.Gen.Virol., 94, 2013
|
|
4F15
 
 | | Molecular basis of infectivity of 2009 pandemic H1N1 influenza A viruses | | Descriptor: | Fab fragment, heavy chain, light chain, ... | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Park, Y.H, Khan, T.G, Lee, J.Y, Kang, S.H, Alam, I. | | Deposit date: | 2012-05-06 | | Release date: | 2013-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Molecular basis of infectivity of 2009 pandemic H1N1 influenza A viruses
To be Published
|
|
4EDB
 
 | | Structures of monomeric hemagglutinin and its complex with an Fab fragment of a neutralizing antibody that binds to H1 subtype influenza viruses: molecular basis of infectivity of 2009 pandemic H1N1 influenza A viruses | | Descriptor: | Hemagglutinin | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Park, Y.H, Khan, T.G, Lee, J.Y, Kang, S.H, Alam, I. | | Deposit date: | 2012-03-27 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into structural diversity of influenza virus haemagglutinin
J.Gen.Virol., 94, 2013
|
|
1Q1Y
 
 | | Crystal Structures of Peptide Deformylase from Staphylococcus aureus Complexed with Actinonin | | Descriptor: | ACTINONIN, Peptide deformylase, ZINC ION | | Authors: | Yoon, H.J, Lee, S.K, Kim, H.L, Kim, H.W, Kim, H.W, Lee, J.Y, Mikami, B, Suh, S.W. | | Deposit date: | 2003-07-23 | | Release date: | 2004-07-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of peptide deformylase from Staphylococcus aureus in complex with actinonin, a naturally occurring antibacterial agent
Proteins, 57, 2004
|
|
7RSJ
 
 | | Structure of the VPS34 kinase domain with compound 14 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-{4-[(7R,8R)-4-oxo-7-(propan-2-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazin-2-yl]pyridin-2-yl}cyclopropanecarboxamide, ... | | Authors: | Hu, D.X, Patel, S, Chen, H, Wang, S, Staben, S, Dimitrova, Y.N, Wallweber, H.A, Lee, J.Y, Chan, G.K.Y, Sneeringer, C.J, Prangley, M.S, Moffat, J.G, Wu, C, Schutt, L.K, Salphati, L, Pang, J, McNamara, E, Huang, H, Chen, Y, Wang, Y, Zhao, W, Lim, J, Murthy, A, Siu, M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | Structure-Based Design of Potent, Selective, and Orally Bioavailable VPS34 Kinase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7RSP
 
 | | Structure of the VPS34 kinase domain with compound 14 | | Descriptor: | (7R,8R)-2-[(3R)-3-methylmorpholin-4-yl]-7-(propan-2-yl)-6,7-dihydropyrazolo[1,5-a]pyrazin-4(5H)-one, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3 | | Authors: | Hu, D.X, Patel, S, Chen, H, Wang, S, Staben, S, Dimitrova, Y.N, Wallweber, H.A, Lee, J.Y, Chan, G.K.Y, Sneeringer, C.J, Prangley, M.S, Moffat, J.G, Wu, C, Schutt, L.K, Salphati, L, Pang, J, McNamara, E, Huang, H, Chen, Y, Wang, Y, Zhao, W, Lim, J, Murthy, A, Siu, M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure-Based Design of Potent, Selective, and Orally Bioavailable VPS34 Kinase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7RSV
 
 | | Structure of the VPS34 kinase domain with compound 5 | | Descriptor: | (5aS,8aR,9S)-2-[(3R)-3-methylmorpholin-4-yl]-5,5a,6,7,8,8a-hexahydro-4H-cyclopenta[e]pyrazolo[1,5-a]pyrazin-4-one, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3, ... | | Authors: | Hu, D.X, Patel, S, Chen, H, Wang, S, Staben, S, Dimitrova, Y.N, Wallweber, H.A, Lee, J.Y, Chan, G.K.Y, Sneeringer, C.J, Prangley, M.S, Moffat, J.G, Wu, C, Schutt, L.K, Salphati, L, Pang, J, McNamara, E, Huang, H, Chen, Y, Wang, Y, Zhao, W, Lim, J, Murthy, A, Siu, M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-Based Design of Potent, Selective, and Orally Bioavailable VPS34 Kinase Inhibitors.
J.Med.Chem., 65, 2022
|
|
1J9L
 
 | | CRYSTAL STRUCTURE OF SURE PROTEIN FROM T.MARITIMA IN COMPLEX WITH VANADATE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, STATIONARY PHASE SURVIVAL PROTEIN, ... | | Authors: | Suh, S.W, Lee, J.Y, Kwak, J.E, Moon, J. | | Deposit date: | 2001-05-28 | | Release date: | 2001-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and functional analysis of the SurE protein identify a novel phosphatase family.
Nat.Struct.Biol., 8, 2001
|
|
1J9J
 
 | | CRYSTAL STRUCTURE ANALYSIS OF SURE PROTEIN FROM T.MARITIMA | | Descriptor: | MAGNESIUM ION, STATIONARY PHASE SURVIVAL PROTEIN, SULFATE ION | | Authors: | Suh, S.W, Lee, J.Y, Kwak, J.E, Moon, J. | | Deposit date: | 2001-05-27 | | Release date: | 2001-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and functional analysis of the SurE protein identify a novel phosphatase family.
Nat.Struct.Biol., 8, 2001
|
|
1J9K
 
 | | CRYSTAL STRUCTURE OF SURE PROTEIN FROM T.MARITIMA IN COMPLEX WITH TUNGSTATE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, STATIONARY PHASE SURVIVAL PROTEIN, ... | | Authors: | Suh, S.W, Lee, J.Y, Kwak, J.E, Moon, J. | | Deposit date: | 2001-05-27 | | Release date: | 2001-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and functional analysis of the SurE protein identify a novel phosphatase family.
Nat.Struct.Biol., 8, 2001
|
|
8CUB
 
 | |
3FS5
 
 | |
1FK2
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH MYRISTIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, MYRISTIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK7
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH RICINOLEIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, NON-SPECIFIC LIPID TRANSFER PROTEIN, RICINOLEIC ACID | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK3
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH PALMITOLEIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN, PALMITOLEIC ACID | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK0
 
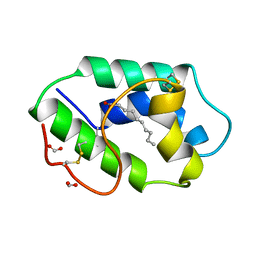 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH CAPRIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | DECANOIC ACID, FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-08 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK5
 
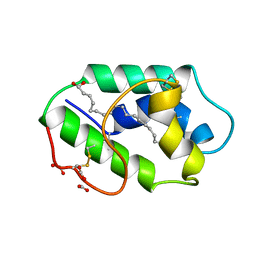 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH OLEIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN, OLEIC ACID | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK4
 
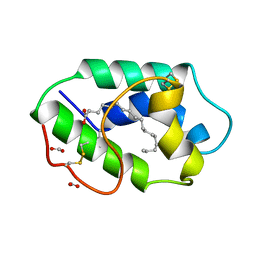 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH STEARIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN, STEARIC ACID | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
2JRV
 
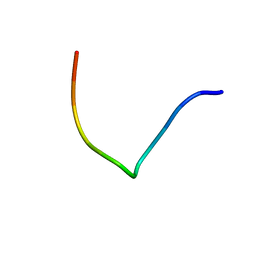 | | The third dimensional structure of mab198-bound pep.1 for autoimmune myasthenia gravis | | Descriptor: | PEPTIDE PEP.1 | | Authors: | Jung, H.H, Yi, H.J, Lee, S.K, Lee, J.Y, Jung, H.J, Yang, S.T, Eu, Y.-J, Im, S.-H, Kim, J.I. | | Deposit date: | 2007-06-29 | | Release date: | 2008-07-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of immunotherapeutic peptides for autoimmune Myasthenia gravis
Biochemistry, 46, 2007
|
|
2JRW
 
 | | Solution structure of Cyclic extended Pep1(Cyc.ext.Pep.1) for autoimmune myasthenia gravis | | Descriptor: | Cyclic extended Pep.1 | | Authors: | Jung, H.H, Yi, H.J, Lee, S.K, Lee, J.Y, Jung, H.J, Yang, S.T, Eu, Y.-J, Im, S.-H, Kim, J.I. | | Deposit date: | 2007-06-29 | | Release date: | 2008-07-01 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of immunotherapeutic peptides for autoimmune Myasthenia gravis
Biochemistry, 46, 2007
|
|
1FK1
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH LAURIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, LAURIC ACID, NON-SPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK6
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH ALPHA-LINOLENIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | ALPHA-LINOLENIC ACID, FORMIC ACID, NON-SPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1MZL
 
 | | MAIZE NONSPECIFIC LIPID TRANSFER PROTEIN | | Descriptor: | MAIZE NONSPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Shin, D.H, Lee, J.Y, Hwang, K.Y, Kim, K.K, Suh, S.W. | | Deposit date: | 1995-01-26 | | Release date: | 1996-08-01 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structure of the non-specific lipid-transfer protein from maize seedlings.
Structure, 3, 1995
|
|
1I3C
 
 | | RESPONSE REGULATOR FOR CYANOBACTERIAL PHYTOCHROME, RCP1 | | Descriptor: | RESPONSE REGULATOR RCP1, SULFATE ION | | Authors: | Im, Y.J, Rho, S.-H, Park, C.-M, Yang, S.-S, Kang, J.-G, Lee, J.Y, Song, P.-S, Eom, S.H. | | Deposit date: | 2001-02-14 | | Release date: | 2002-03-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a cyanobacterial phytochrome response regulator.
Protein Sci., 11, 2002
|
|
1NNE
 
 | | Crystal Structure of the MutS-ADPBeF3-DNA complex | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*GP*CP*GP*AP*CP*GP*CP*TP*AP*GP*CP*GP*TP*GP*CP*GP*GP*CP*TP*CP*GP*TP*C)-3', 5'-D(P*GP*GP*AP*CP*GP*AP*GP*CP*CP*GP*CP*CP*GP*CP*TP*AP*GP*CP*GP*TP*CP*G)-3', ... | | Authors: | Alani, E, Lee, J.Y, Schofield, M.J, Kijas, A.W, Hsieh, P, Yang, W. | | Deposit date: | 2003-01-13 | | Release date: | 2003-05-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structure and biochemical analysis of the MutS-ADP-Beryllium Fluoride complex
suggests a conserved mechanism for ATP interactions in mismatch repair
J.Biol.Chem., 278, 2003
|
|
