5H78
 
 | | Crystal structure of the PKA-DHR14 fusion protein | | Descriptor: | cAMP-dependent protein kinase type II-alpha regulatory subunit,DHR14 | | Authors: | Youn, S.J, Kwon, N.Y, Lee, J.H, Kim, J.H, Lee, H, Lee, J.O. | | Deposit date: | 2016-11-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Construction of novel repeat proteins with rigid and predictable structures using a shared helix method.
Sci Rep, 7, 2017
|
|
5H7C
 
 | | Crystal structure of a repeat protein with two Protein A-DHR14 repeat modules | | Descriptor: | Immunoglobulin G-binding protein A, DHR14 | | Authors: | Youn, S.J, Kwon, N.Y, Lee, J.H, Kim, J.H, Lee, H, Lee, J.O. | | Deposit date: | 2016-11-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Construction of novel repeat proteins with rigid and predictable structures using a shared helix method.
Sci Rep, 7, 2017
|
|
5H77
 
 | | Crystal structure of the PKA-protein A fusion protein | | Descriptor: | cAMP-dependent protein kinase type II-alpha regulatory subunit,Immunoglobulin G-binding protein A | | Authors: | Youn, S.J, Kwon, N.Y, Lee, J.H, Kim, J.H, Lee, H, Lee, J.O. | | Deposit date: | 2016-11-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.197 Å) | | Cite: | Construction of novel repeat proteins with rigid and predictable structures using a shared helix method.
Sci Rep, 7, 2017
|
|
5H79
 
 | | Crystal structure of a repeat protein with three Protein A repeat module | | Descriptor: | Immunoglobulin G-binding protein A | | Authors: | Youn, S.J, Kwon, N.Y, Lee, J.H, Kim, J.H, Lee, H, Lee, J.O. | | Deposit date: | 2016-11-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Construction of novel repeat proteins with rigid and predictable structures using a shared helix method.
Sci Rep, 7, 2017
|
|
5H7B
 
 | | Crystal structure of a repeat protein with five Protein A repeat modules | | Descriptor: | Immunoglobulin G-binding protein A | | Authors: | Youn, S.J, Kwon, N.Y, Lee, J.H, Kim, J.H, Lee, H, Lee, J.O. | | Deposit date: | 2016-11-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Construction of novel repeat proteins with rigid and predictable structures using a shared helix method.
Sci Rep, 7, 2017
|
|
5H7A
 
 | | Crystal structure of a repeat protein with four Protein A repeat module | | Descriptor: | Immunoglobulin G-binding protein A | | Authors: | Youn, S.J, Kwon, N.Y, Lee, J.H, Kim, J.H, Lee, H, Lee, J.O. | | Deposit date: | 2016-11-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Construction of novel repeat proteins with rigid and predictable structures using a shared helix method.
Sci Rep, 7, 2017
|
|
5H76
 
 | | Crystal structure of the DARPin-Protein A fusion protein | | Descriptor: | DARPin,Immunoglobulin G-binding protein A | | Authors: | Youn, S.J, Kwon, N.Y, Lee, J.H, Kim, J.H, Lee, H, Lee, J.O. | | Deposit date: | 2016-11-17 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Construction of novel repeat proteins with rigid and predictable structures using a shared helix method.
Sci Rep, 7, 2017
|
|
8WFC
 
 | |
7X99
 
 | |
7XJT
 
 | |
4F15
 
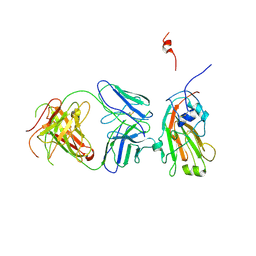 | | Molecular basis of infectivity of 2009 pandemic H1N1 influenza A viruses | | Descriptor: | Fab fragment, heavy chain, light chain, ... | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Park, Y.H, Khan, T.G, Lee, J.Y, Kang, S.H, Alam, I. | | Deposit date: | 2012-05-06 | | Release date: | 2013-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Molecular basis of infectivity of 2009 pandemic H1N1 influenza A viruses
To be Published
|
|
5H1Z
 
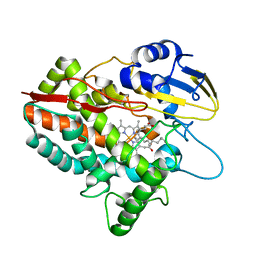 | | CYP153D17 from Sphingomonas sp. PAMC 26605 | | Descriptor: | DODECANE, PROTOPORPHYRIN IX CONTAINING FE, putative CYP alkane hydroxylase CYP153D17 | | Authors: | Lee, C.W, Lee, J.H. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of a Putative Cytochrome P450 Alkane Hydroxylase (CYP153D17) from Sphingomonas sp. PAMC 26605 and Its Conformational Substrate Binding
Int J Mol Sci, 17, 2016
|
|
7WG3
 
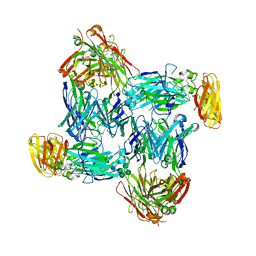 | | Structural basis of interleukin-17B receptor in complex with a neutralizing antibody D9 for guiding humanization and affinity maturation for cancer therapy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D9 Fab, IL17RB protein, ... | | Authors: | Lee, W.H, Chen, X.R, Liu, I.J, Lee, J.H, Hu, C.M, Wu, H.C, Wang, S.K, Lee, W.H, Ma, C. | | Deposit date: | 2021-12-28 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural basis of interleukin-17B receptor in complex with a neutralizing antibody for guiding humanization and affinity maturation.
Cell Rep, 41, 2022
|
|
6X6P
 
 | | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Herrera, N.G, Morano, N.C, Celikgil, A, Georgiev, G.I, Malonis, R, Lee, J.H, Tong, K, Vergnolle, O, Massimi, A, Yen, L.Y, Noble, A.J, Kopylov, M, Bonanno, J.B, Garrett-Thompson, S.C, Hayes, D.B, Brenowitz, M, Garforth, S.J, Eng, E.T, Lai, J.R, Almo, S.C. | | Deposit date: | 2020-05-28 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis.
Biorxiv, 2020
|
|
5IT1
 
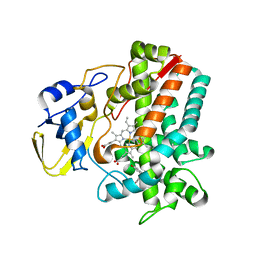 | | Streptomyces peucetius CYP105P2 complex with biphenyl compound | | Descriptor: | 4,4'-PROPANE-2,2-DIYLDIPHENOL, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 | | Authors: | Lee, C.W, Lee, J.H. | | Deposit date: | 2016-03-16 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Cytochrome P450 (CYP105P2) from Streptomyces peucetius and Its Conformational Changes in Response to Substrate Binding
Int J Mol Sci, 17, 2016
|
|
6J4N
 
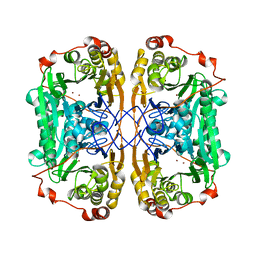 | | Structure of papua new guinea MBL-1(PNGM-1) native | | Descriptor: | Metallo-beta-lactamases PNGM-1, ZINC ION | | Authors: | Hong, M.K, Park, K.S, Jeon, J.H, Lee, J.H, Park, Y.S, Lee, S.H, Kang, L.W. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The novel metallo-beta-lactamase PNGM-1 from a deep-sea sediment metagenome: crystallization and X-ray crystallographic analysis.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7YC0
 
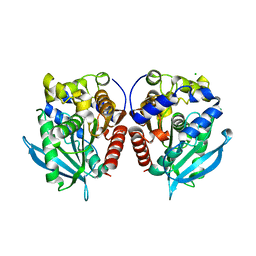 | | Acetylesterase (LgEstI) W.T. | | Descriptor: | ACETATE ION, Alpha/beta hydrolase, CHLORIDE ION | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2022-06-30 | | Release date: | 2023-06-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and biochemical analysis of acetylesterase (LgEstI) from Lactococcus garvieae.
Plos One, 18, 2023
|
|
7YC4
 
 | | Acetylesterase (LgEstI) F207A | | Descriptor: | Alpha/beta hydrolase | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2022-06-30 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and biochemical analysis of acetylesterase (LgEstI) from Lactococcus garvieae.
Plos One, 18, 2023
|
|
7VPF
 
 | |
8ILJ
 
 | |
8HEA
 
 | | Esterase2 (EaEst2) from Exiguobacterium antarcticum | | Descriptor: | Thermostable carboxylesterase Est30 | | Authors: | Hwang, J, Lee, J.H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and Biochemical Insights into Bis(2-hydroxyethyl) Terephthalate Degrading Carboxylesterase Isolated from Psychrotrophic Bacterium Exiguobacterium antarcticum.
Int J Mol Sci, 24, 2023
|
|
7NFX
 
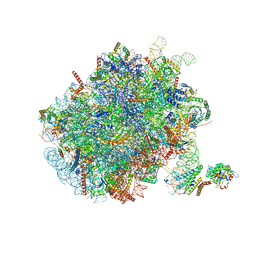 | | Mammalian ribosome nascent chain complex with SRP and SRP receptor in early state A | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Jomaa, A, Lee, J.H, Shan, S, Ban, N. | | Deposit date: | 2021-02-08 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Receptor compaction and GTPase rearrangement drive SRP-mediated cotranslational protein translocation into the ER.
Sci Adv, 7, 2021
|
|
5D9V
 
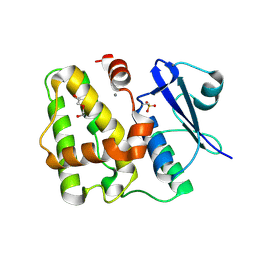 | |
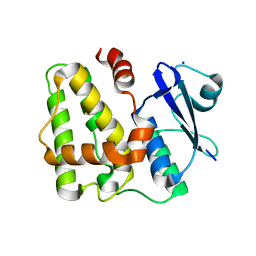
5D9T
 
 | |
5D9X
 
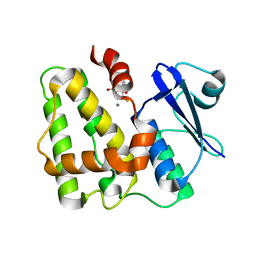 | | Dehydroascorbate reductase complexed with GSH | | Descriptor: | CALCIUM ION, Dehydroascorbate reductase, GLUTATHIONE | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2015-08-19 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural understanding of the recycling of oxidized ascorbate by dehydroascorbate reductase (OsDHAR) from Oryza sativa L. japonica
Sci Rep, 6, 2016
|
|
