3VF0
 
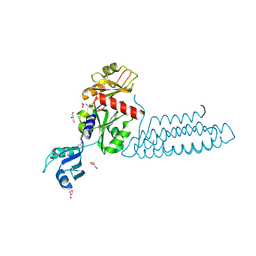 | | Raver1 in complex with metavinculin L954 deletion mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Ribonucleoprotein PTB-binding 1, ... | | Authors: | Lee, J.H, Vonrhein, C, Bricogne, G, Izard, T. | | Deposit date: | 2012-01-09 | | Release date: | 2012-07-25 | | Last modified: | 2012-09-19 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | The Metavinculin Tail Domain Directs Constitutive Interactions with Raver1 and vinculin RNA.
J.Mol.Biol., 422, 2012
|
|
2QZX
 
 | | Secreted aspartic proteinase (Sap) 5 from Candida albicans | | Descriptor: | Candidapepsin-5, Pepstatin | | Authors: | Lee, J.H, Ruge, E, Borelli, C, Maskos, K, Huber, R. | | Deposit date: | 2007-08-17 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structures of Sap1 and Sap5: Structural comparison of the secreted aspartic proteinases from Candida albicans.
Proteins, 72, 2008
|
|
5XGW
 
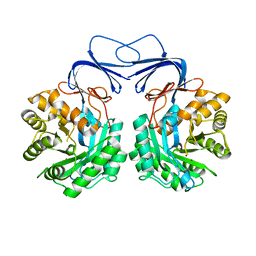 | |
5XGX
 
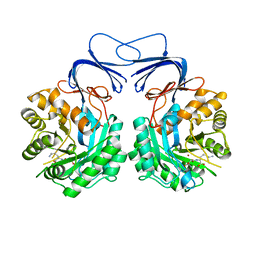 | | Crystal structure of colwellia psychrerythraea strain 34H isoaspartyl dipeptidase E80Q mutant complexed with beta-isoaspartyl lysine | | Descriptor: | D-ASPARTIC ACID, D-LYSINE, Isoaspartyl dipeptidase, ... | | Authors: | Lee, J.H, Lee, C.W, Park, S.H. | | Deposit date: | 2017-04-18 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure and functional characterization of an isoaspartyl dipeptidase (CpsIadA) from Colwellia psychrerythraea strain 34H.
PLoS ONE, 12, 2017
|
|
5Z2E
 
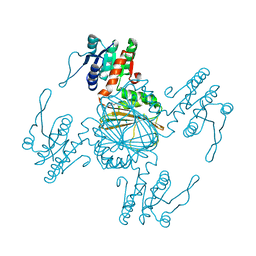 | |
5Z2F
 
 | | NADPH/PDA bound Dihydrodipicolinate reductase from Paenisporosarcina sp. TG-14 | | Descriptor: | Dihydrodipicolinate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYRIDINE-2,6-DICARBOXYLIC ACID | | Authors: | Lee, J.H, Lee, C.W, Park, S. | | Deposit date: | 2018-01-02 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of dihydrodipicolinate reductase (PaDHDPR) from Paenisporosarcina sp. TG-14: structural basis for NADPH preference as a cofactor
Sci Rep, 8, 2018
|
|
5Z2D
 
 | |
5YL7
 
 | | Proteases from Pseudoalteromonas arctica PAMC 21717 (Pro21717) | | Descriptor: | CALCIUM ION, Copurified unknown peptide, Pseudoalteromonas arctica PAMC 21717 | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2017-10-17 | | Release date: | 2018-01-31 | | Last modified: | 2018-09-12 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of a cold-active protease (Pro21717) from the psychrophilic bacterium, Pseudoalteromonas arctica PAMC 21717, at 1.4 angstrom resolution: Structural adaptations to cold and functional analysis of a laundry detergent enzyme
PLoS ONE, 13, 2018
|
|
5WQ0
 
 | | Receiver domain of Spo0A from Paenisporosarcina sp. TG-14 | | Descriptor: | MAGNESIUM ION, Stage 0 sporulation protein | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2016-11-22 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Crystal structure of the inactive state of the receiver domain of Spo0A from Paenisporosarcina sp. TG-14, a psychrophilic bacterium isolated from an Antarctic glacier
J. Microbiol., 55, 2017
|
|
5BVT
 
 | | Palmitate-bound pFABP5 | | Descriptor: | Epidermal fatty acid-binding protein, PALMITOLEIC ACID | | Authors: | Lee, J.H, Lee, C.W, Do, H. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis for the ligand-binding specificity of fatty acid-binding proteins (pFABP4 and pFABP5) in gentoo penguin
Biochem.Biophys.Res.Commun., 465, 2015
|
|
5BVS
 
 | | Linoleate-bound pFABP4 | | Descriptor: | Fatty acid-binding protein, LINOLEIC ACID | | Authors: | Lee, J.H, Lee, C.W, Do, H. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the ligand-binding specificity of fatty acid-binding proteins (pFABP4 and pFABP5) in gentoo penguin
Biochem.Biophys.Res.Commun., 465, 2015
|
|
5BVQ
 
 | | Ligand-unbound pFABP4 | | Descriptor: | fatty acid-binding protein | | Authors: | Lee, J.H, Lee, C.W, Do, H. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the ligand-binding specificity of fatty acid-binding proteins (pFABP4 and pFABP5) in gentoo penguin
Biochem.Biophys.Res.Commun., 465, 2015
|
|
6JQS
 
 | | Structure of Transcription factor, GerE | | Descriptor: | DNA-binding response regulator | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2019-04-01 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a transcription factor, GerE (PaGerE), from spore-forming bacterium Paenisporosarcina sp. TG-14.
Biochem.Biophys.Res.Commun., 513, 2019
|
|
5XWB
 
 | | Crystal Structure of 5-Enolpyruvulshikimate-3-phosphate Synthase from a Psychrophilic Bacterium, Colwellia psychrerythraea | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase | | Authors: | Lee, J.H, Kim, H.J, Choi, J.M, Kim, D.-W, Seo, Y.-S. | | Deposit date: | 2017-06-29 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of 5-enolpyruvylshikimate-3-phosphate synthase from a psychrophilic bacterium, Colwellia psychrerythraea 34H.
Biochem. Biophys. Res. Commun., 492, 2017
|
|
6IFH
 
 | | Unphosphorylated Spo0F from Paenisporosarcina sp. TG-14 | | Descriptor: | MAGNESIUM ION, Sporulation initiation phosphotransferase F | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of unphosphorylated Spo0F from Paenisporosarcina sp. TG-14, a psychrophilic bacterium isolated from an Antarctic glacier
Biodesign, 6(4), 2019
|
|
4LQ8
 
 | |
5BY2
 
 | |
6INT
 
 | | xylose isomerase from Paenibacillus sp. R4 | | Descriptor: | CALCIUM ION, Xylose isomerase | | Authors: | Lee, J.H, Lee, C.W, Park, S. | | Deposit date: | 2018-10-26 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.942 Å) | | Cite: | Crystal Structure and Functional Characterization of a Xylose Isomerase (PbXI) from the Psychrophilic Soil Microorganism, Paenibacillus sp.
J. Microbiol. Biotechnol., 29, 2019
|
|
6IYM
 
 | |
2FB9
 
 | |
2FJK
 
 | | Crystal structure of Fructose-1,6-Bisphosphate Aldolase in Thermus caldophilus | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-bisphosphate aldolase | | Authors: | Lee, J.H, Im, Y.J, Rho, S.-H, Kim, M.-K, Kang, G.B, Eom, S.H. | | Deposit date: | 2006-01-03 | | Release date: | 2006-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Stereoselectivity of fructose-1,6-bisphosphate aldolase in Thermus caldophilus
Biochem.Biophys.Res.Commun., 347, 2006
|
|
7YLK
 
 | | Myoglobin containing Ir complex | | Descriptor: | Myoglobin, SULFATE ION, delta-{1-([2,2'-bipyridin]-5-ylmethyl)pyrrolidine-2,5-dione}bis[2-(2,4-difluorophenyl)pyridine)]iridium(III), ... | | Authors: | Lee, J.H, Song, W.J. | | Deposit date: | 2022-07-26 | | Release date: | 2023-03-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Photocatalytic C-O Coupling Enzymes That Operate via Intramolecular Electron Transfer.
J.Am.Chem.Soc., 145, 2023
|
|
5H3H
 
 | | Esterase (EaEST) from Exiguobacterium antarcticum | | Descriptor: | Abhydrolase domain-containing protein, ETHANEPEROXOIC ACID | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2016-10-24 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure and Functional Characterization of an Esterase (EaEST) from Exiguobacterium antarcticum.
Plos One, 12, 2017
|
|
7C4X
 
 | |
2B8I
 
 | | Crystal Structure and Functional Studies Reveal that PAS Factor from Vibrio vulnificus is a Novel Member of the Saposin-Fold Family | | Descriptor: | PAS factor | | Authors: | Lee, J.H, Yang, S.T, Rho, S.H, Im, Y.J, Kim, S.Y, Kim, Y.R, Kim, M.K, Kang, G.B, Kim, J.I, Rhee, J.H, Eom, S.H. | | Deposit date: | 2005-10-07 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and functional studies reveal that PAS factor from Vibrio vulnificus is a novel member of the saposin-fold family
J.Mol.Biol., 355, 2006
|
|
