6Z6F
 
 | | HDAC-PC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
6Z6H
 
 | | HDAC-DC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (8.55 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
6Z6P
 
 | | HDAC-PC-Nuc | | Descriptor: | DNA (145-MER), HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
6Z6O
 
 | | HDAC-TC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
1QL5
 
 | | DNA DECAMER DUPLEX CONTAINING T5-T6 PHOTOADDUCT | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*TP*+TP*AP*CP*GP*C)- 3'), DNA (5'-D(*GP*CP*GP*TP*TP*AP*TP*GP*CP*G)-3') | | Authors: | Lee, J.-H, Hwang, G.-S, Choi, B.-S. | | Deposit date: | 1999-08-24 | | Release date: | 2000-04-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a DNA Decamer Duplex Containing the 3' T.T Base Pair of the Cis-Syn Cyclobutane Pyrimidine Dimer: Implication for the Mutagenic Property of the Cis-Syn Dimer.
Nucleic Acids Res., 28, 2000
|
|
1QKG
 
 | | DNA DECAMER DUPLEX CONTAINING T-T DEWAR PHOTOPRODUCT | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*(HYD)TP*+TP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*AP*TP*GP*CP*G)-3') | | Authors: | Lee, J.-H, Bae, S.-H, Choi, Y.-J, Choi, B.-S. | | Deposit date: | 1999-07-20 | | Release date: | 2000-05-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Dewar Photoproduct of Thymidylyl(3'-->5')-Thymidine (Dewar Product) Exhibits Mutagenic Behavior in Accordance with the "A Rule".
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
4OWI
 
 | | peptide structure | | Descriptor: | p53LZ2 | | Authors: | Lee, J.-H. | | Deposit date: | 2014-02-02 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.202 Å) | | Cite: | Protein grafting of p53TAD onto a leucine zipper scaffold generates a potent HDM dual inhibitor.
Nat Commun, 5, 2014
|
|
8HMW
 
 | |
1X3Z
 
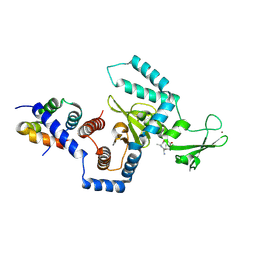 | | Structure of a peptide:N-glycanase-Rad23 complex | | Descriptor: | UV excision repair protein RAD23, ZINC ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, ... | | Authors: | Lee, J.-H, Choi, J.M, Lee, C, Yi, K.J, Cho, Y. | | Deposit date: | 2005-05-11 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a peptide:N-glycanase-Rad23 complex: insight into the deglycosylation for denatured glycoproteins.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1X3W
 
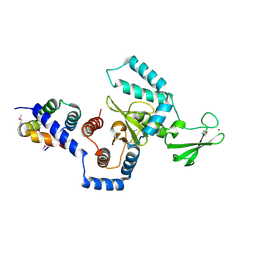 | | Structure of a peptide:N-glycanase-Rad23 complex | | Descriptor: | UV excision repair protein RAD23, ZINC ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, ... | | Authors: | Lee, J.-H, Choi, J.M, Lee, C, Yi, K.J, Cho, Y. | | Deposit date: | 2005-05-11 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a peptide:N-glycanase-Rad23 complex: insight into the deglycosylation for denatured glycoproteins.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
8OM9
 
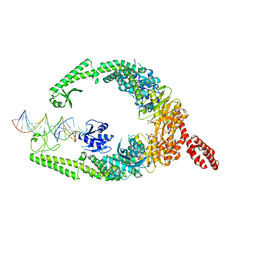 | | MutSbeta bound to (CAG)2 DNA (open form) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein Msh2, DNA mismatch repair protein Msh3, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | MutSbeta bound to (CAG)2 DNA (open form)
To Be Published
|
|
8OLX
 
 | | MutSbeta bound to (CAG)2 DNA (canonical form) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (25-MER), DNA mismatch repair protein Msh2, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2023-03-30 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | MutSbeta bound to (CAG)2 DNA (canonical form)
To Be Published
|
|
8OMO
 
 | | DNA-unbound MutSbeta-ATP complex (bent clamp form) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA mismatch repair protein Msh2, DNA mismatch repair protein Msh3, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | DNA-unbound MutSbeta-ATP complex (bent clamp form)
To Be Published
|
|
8OM5
 
 | | DNA-free open form of MutSbeta | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein Msh2, DNA mismatch repair protein Msh3, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | DNA-free open form of MutSbeta
To Be Published
|
|
8OMA
 
 | | MutSbeta bound to 61bp homoduplex DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA mismatch repair protein Msh2, DNA mismatch repair protein Msh3, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | MutSbeta bound to 61bp homoduplex DNA
To Be Published
|
|
8OMQ
 
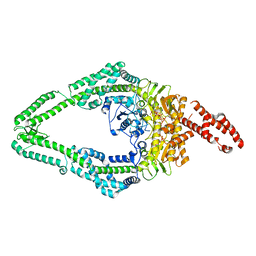 | | DNA-unbound MutSbeta-ATP complex (straight clamp form) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA mismatch repair protein Msh2, DNA mismatch repair protein Msh3, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | DNA-unbound MutSbeta-ATP complex (straight clamp form)
To Be Published
|
|
1CFL
 
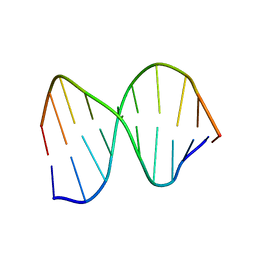 | | DNA DECAMER DUPLEX CONTAINING T5-T6 PHOTOADDUCT | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*(64T)P*TP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*AP*TP*GP*CP*G)-3') | | Authors: | Lee, J.-H, Hwang, G.-S, Choi, B.-S. | | Deposit date: | 1999-03-19 | | Release date: | 1999-05-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA decamer duplex containing the stable 3' T.G base pair of the pyrimidine(6-4)pyrimidone photoproduct [(6-4) adduct]: implications for the highly specific 3' T --> C transition of the (6-4) adduct.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
4LCT
 
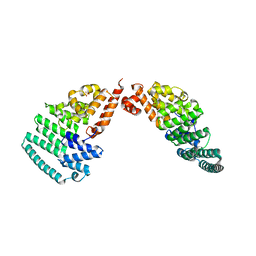 | | Crystal Structure and Versatile Functional Roles of the COP9 Signalosome Subunit 1 | | Descriptor: | COP9 signalosome complex subunit 1, SULFATE ION | | Authors: | Lee, J.-H, Yi, L, Li, J, Schweitzer, K, Borgmann, M, Naumann, M, Wu, H. | | Deposit date: | 2013-06-23 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and versatile functional roles of the COP9 signalosome subunit 1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1MNL
 
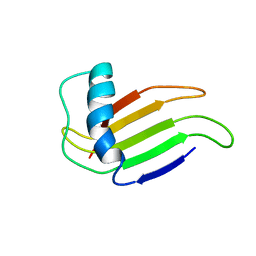 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF A SWEET PROTEIN SINGLE-CHAIN MONELLIN (SCM) DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND DYNAMICAL SIMULATED ANNEALING CALCULATIONS, 21 STRUCTURES | | Descriptor: | MONELLIN | | Authors: | Lee, S.-Y, Lee, J.-H, Chang, H.-J, Jo, J.-M, Jung, J.-W, Lee, W. | | Deposit date: | 1998-08-06 | | Release date: | 1999-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a sweet protein single-chain monellin determined by nuclear magnetic resonance and dynamical simulated annealing calculations.
Biochemistry, 38, 1999
|
|
4XCS
 
 | | Human peroxiredoxin-1 C83S mutant | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, GLYCEROL, Peroxiredoxin-1 | | Authors: | Cho, K.J, Lee, J.-H, Khan, T.G, Park, Y, Cho, A, Chang, T.-S, Kim, K.H. | | Deposit date: | 2014-12-18 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Dimeric Human Peroxiredoxin-1 C83S Mutant
Bull.Korean Chem.Soc., 36, 2015
|
|
1JO7
 
 | | Solution Structure of Influenza A Virus Promoter | | Descriptor: | Influenza A virus promoter RNA | | Authors: | Bae, S.-H, Cheong, H.-K, Lee, J.-H, Cheong, C, Kainosho, M, Choi, B.-S. | | Deposit date: | 2001-07-27 | | Release date: | 2001-09-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural features of an influenza virus promoter and their implications for viral RNA synthesis.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4ES0
 
 | | X-ray structure of WDR5-SETd1b Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase SETD1B, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-21 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.817 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4ERY
 
 | | X-ray structure of WDR5-MLL3 Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase MLL3, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-21 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4ERQ
 
 | | X-ray structure of WDR5-MLL2 Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase MLL2, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4EWR
 
 | | X-ray structure of WDR5-SETd1a Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase SETD1A, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-27 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
