8UEC
 
 | |
3JBA
 
 | | The U4 antibody epitope on human papillomavirus 16 identified by cryo-EM | | Descriptor: | H16.U4 antibody heavy chain, H16.U4 antibody light chain, Major capsid protein L1 | | Authors: | Guan, J, Bywaters, S.M, Brendle, S.A, Lee, H, Ashley, R.E, Christensen, N.D, Hafenstein, S. | | Deposit date: | 2015-08-11 | | Release date: | 2015-10-07 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | The U4 Antibody Epitope on Human Papillomavirus 16 Identified by Cryo-electron Microscopy.
J.Virol., 89, 2015
|
|
1TWL
 
 | | Inorganic pyrophosphatase from Pyrococcus furiosus Pfu-264096-001 | | Descriptor: | Inorganic pyrophosphatase | | Authors: | Zhou, W, Tempel, W, Liu, Z.-J, Chen, L, Clancy Kelley, L.-L, Dillard, B.D, Hopkins, R.C, Arendall III, W.B, Rose, J.P, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, J.S, Richardson, D.C, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-07-01 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Inorganic pyrophosphatase from Pyrococcus furiosus Pfu-264096-001
To be published
|
|
2KYB
 
 | | Solution structure of CpR82G from Clostridium perfringens. North East Structural Genomics Consortium Target CpR82g | | Descriptor: | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase domain protein, possible enterotoxin | | Authors: | Mobley, C.K, Lee, H, Lee, D, Ciccosanti, C, Janjua, H, Acton, T.B, Xiao, R, Everrett, J.K, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of CpR82G
To be Published
|
|
1JOM
 
 | | THE CRYSTAL STRUCTURE OF THE BINARY COMPLEX BETWEEN FOLINIC ACID (LEUCOVORIN) AND E. COLI DIHYDROFOLATE REDUCTASE | | Descriptor: | CALCIUM ION, CHLORIDE ION, DIHYDROFOLATE REDUCTASE, ... | | Authors: | Reyes, V.M, Lee, H, Kraut, J. | | Deposit date: | 1996-02-25 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Escherichia coli dihydrofolate reductase complexed with 5-formyltetrahydrofolate (folinic acid) in two space groups: evidence for enolization of pteridine O4.
Biochemistry, 35, 1996
|
|
6DIZ
 
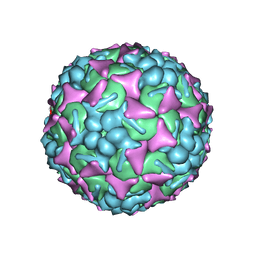 | | EV-A71 strain 11316 complexed with tryptophan dendrimer MADAL_0385 | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Sun, L, Lee, H, Thibaut, H.J, Rivero-Buceta, E, Martinez-Gualda, B, Delang, L, Leyssen, P, Gago, F, San-Felix, A, Hafenstein, S, Mirabelli, C, Neyts, J. | | Deposit date: | 2018-05-24 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Viral engagement with host (co-)receptors blocked by a novel class of tryptophan dendrimers that targets the 5-fold-axis of the enterovirus-A71 capsid.
Plos Pathog., 15, 2019
|
|
5T1V
 
 | | Crystal structure of Zika virus NS2B-NS3 protease in apo-form. | | Descriptor: | NS2B-NS3 protease,NS2B-NS3 protease | | Authors: | Nocadello, S, Light, S.H, Minasov, G, Shuvalova, L, Cardona-Correa, A.A, Ojeda, I, Vargas, J, Johnson, M.E, Lee, H, Anderson, W.F, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-22 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of Zika virus NS2B-NS3 protease in apo-form.
To Be Published
|
|
5TXG
 
 | | Crystal structure of the Zika virus NS3 helicase. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NS3 helicase, POTASSIUM ION | | Authors: | Nocadello, S, Light, S.H, Minasov, G, Shuvalova, L, Cardona-Correa, A.A, Ojeda, I, Vargas, J, Johnson, M.E, Lee, H, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-16 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the Zika virus NS3 helicase.
To be published
|
|
2LTT
 
 | | Solution NMR Structure of YdbC:dT19G1 complex. Northeast Structural Genomics Consortium (NESG) Target KR150 | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Putative uncharacterized protein ydbC | | Authors: | Rossi, P, Barbieri, C.M, Aramini, J.A, Bini, E, Lee, H, Janjua, H, Ciccosanti, C, Wang, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-31 | | Release date: | 2012-06-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of apo- and ssDNA-bound YdbC from Lactococcus lactis uncover the function of protein domain family DUF2128 and expand the single-stranded DNA-binding domain proteome.
Nucleic Acids Res., 41, 2013
|
|
2LDU
 
 | | Solution NMR Structure of Heat shock factor protein 1 DNA binding domain from homo sapiens, Northeast Structural Genomics Consortium Target HR3023C | | Descriptor: | Heat shock factor protein 1 | | Authors: | Liu, G, Xiao, R, Ciccosanti, C, Janjua, H, Acton, T.B, Lee, H, Wang, H.B, Huang, Y.B, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-01 | | Release date: | 2011-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target HR3023C
To be Published
|
|
4ZSW
 
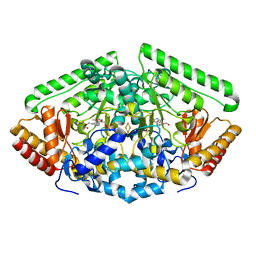 | | Pig Brain GABA-AT inactivated by (E)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic acid | | Descriptor: | (1S)-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopent-3-ene-1,3-dicarboxylic acid, 4-aminobutyrate aminotransferase, mitochondrial, ... | | Authors: | Wu, R, Lee, H, Le, H.V, Doud, E, Sanishvili, R, Compton, P, Kelleher, N.L, Silverman, R.B, Liu, D. | | Deposit date: | 2015-05-14 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Inactivation of GABA Aminotransferase by (E)- and (Z)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic Acid.
Acs Chem.Biol., 10, 2015
|
|
4ZSY
 
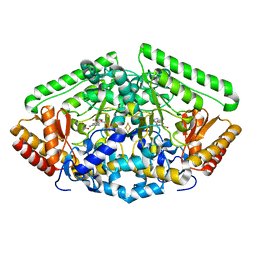 | | Pig Brain GABA-AT inactivated by (Z)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic acid. | | Descriptor: | (1S)-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopent-3-ene-1,3-dicarboxylic acid, 4-aminobutyrate aminotransferase, mitochondrial, ... | | Authors: | Wu, R, Lee, H, Le, H.V, Doud, E, Sanishvili, R, Compton, P, Kelleher, N.L, Silverman, R.B, Liu, D. | | Deposit date: | 2015-05-14 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Inactivation of GABA Aminotransferase by (E)- and (Z)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic Acid.
Acs Chem.Biol., 10, 2015
|
|
2LTD
 
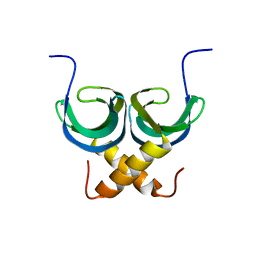 | | Solution NMR Structure of apo YdbC from Lactococcus lactis, Northeast Structural Genomics Consortium (NESG) Target KR150 | | Descriptor: | Uncharacterized protein ydbC | | Authors: | Rossi, P, Barbieri, C.M, Aramini, J.M, Bini, E, Lee, H, Janjua, H, Ciccosanti, C, Wang, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-16 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of apo- and ssDNA-bound YdbC from Lactococcus lactis uncover the function of protein domain family DUF2128 and expand the single-stranded DNA-binding domain proteome.
Nucleic Acids Res., 41, 2013
|
|
2KL3
 
 | | Solution NMR structure of the Rhodanese-like domain from Anabaena sp Alr3790 protein. Northeast Structural Genomics Consortium Target NsR437A | | Descriptor: | Alr3790 protein | | Authors: | Eletsky, A, Belote, R.L, Ciccosanti, C, Janjua, H, Nair, R, Rost, B, Swapna, G.V.T, Acton, T.B, Xiao, R, Everett, J.K, Lee, H, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the Rhodanese-like domain from Anabaena sp Alr3790 protein. Northeast Structural Genomics Consortium Target NsR437A
To be Published
|
|
2KCT
 
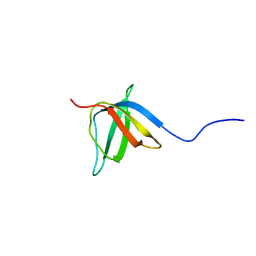 | | Solution nmr structure of the ob-fold domain of heme chaperone ccme from desulfovibrio vulgaris. northeast structural genomics target dvr115g. | | Descriptor: | Cytochrome c-type biogenesis protein CcmE | | Authors: | Aramini, J.M, Rossi, P, Lee, H, Lemak, A, Wang, H, Foote, E.L, Jiang, M, Xiao, R, Nair, R, Swapna, G.V.T, Acton, T.B, Rost, B, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-29 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution nmr structure of the ob-fold domain of heme chaperone
ccme from desulfovibrio vulgaris. northeast structural genomics target dvr115g.
To be Published
|
|
2LCI
 
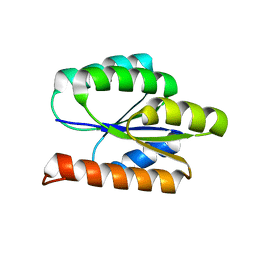 | | Solution NMR Structure of DE NOVO DESIGNED PROTEIN, P-LOOP NTPASE FOLD, Northeast Structural Genomics Consortium Target OR36 (CASD Target) | | Descriptor: | Protein OR36 | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Janjua, H, Ciccosanti, C, Lee, H, Acton, T.B, Everett, J, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-04-29 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target OR36
To be Published
|
|
1YMZ
 
 | | CC45, An Artificial WW Domain Designed Using Statistical Coupling Analysis | | Descriptor: | CC45 | | Authors: | Socolich, M, Lockless, S.W, Russ, W.P, Lee, H, Gardner, K.H, Ranganathan, R. | | Deposit date: | 2005-01-22 | | Release date: | 2005-09-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Evolutionary information for specifying a protein fold.
Nature, 437, 2005
|
|
3FCA
 
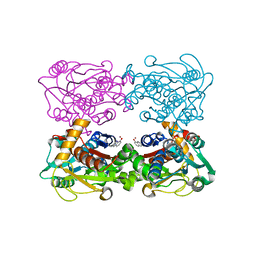 | | Genetic Incorporation of a Metal-ion Chelating Amino Acid into proteins as biophysical probe | | Descriptor: | Cysteine synthase, ZINC ION | | Authors: | Wang, F, Lee, H, Spraggon, G, Schultz, P.G. | | Deposit date: | 2008-11-21 | | Release date: | 2009-02-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Genetic incorporation of a metal-ion chelating amino acid into proteins as a biophysical probe.
J.Am.Chem.Soc., 131, 2009
|
|
2LPK
 
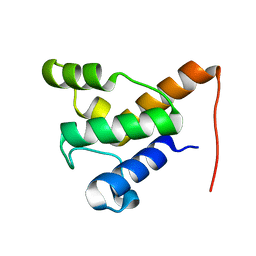 | | Solution NMR of the specialized apo-acyl carrier protein (RPA2022) from Rhodopseudomonas palustris, Northeast Structural Genomics Consortium Target RpR324 | | Descriptor: | Specialized acyl carrier protein | | Authors: | Ramelot, T.A, Rossi, P, Yang, Y, Lee, H, Ertekin, A, Wang, H, Ciccosanti, C, Kohan, E, Maglaqui, M, Janjua, H, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-02-14 | | Release date: | 2012-02-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a specialized acyl carrier protein essential for lipid A biosynthesis with very long-chain fatty acids in open and closed conformations.
Biochemistry, 51, 2012
|
|
2LNU
 
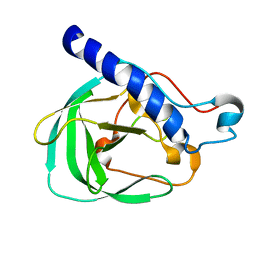 | | Solution NMR Structure of the uncharacterized protein from gene locus rrnAC0354 of Haloarcula marismortui, Northeast Structural Genomics Consortium Target HmR11 | | Descriptor: | Uncharacterized protein | | Authors: | Rossi, P, Liu, G, Lange, O.F, Lee, H, Janjua, H, Ciccosanti, C, Wang, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-01-05 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2L7K
 
 | | Solution NMR Structure of protein CD1104.2 from Clostridium difficile, Northeast Structural Genomics Consortium Target CfR130 | | Descriptor: | Uncharacterized protein | | Authors: | Pulavarti, S.V.S.R.K, Eletsky, A, Mills, J.L, Sukumaran, D.K, Wang, H, Ciccosanti, C, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Lee, H, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-13 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of CfR130 from Clostridium difficile, Northeast Structural Genomics Consortium Target CfR130
To be Published
|
|
4RDI
 
 | | Crystal structure of E. coli tRNA N6-threonylcarbamoyladenosine dehydratase, TcdA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, tRNA threonylcarbamoyladenosine dehydratase | | Authors: | Park, S.Y, Kim, S, Lee, H. | | Deposit date: | 2014-09-19 | | Release date: | 2015-08-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Structure of Escherichia coli TcdA (Also Known As CsdL) Reveals a Novel Topology and Provides Insight into the tRNA Binding Surface Required for N(6)-Threonylcarbamoyladenosine Dehydratase Activity.
J.Mol.Biol., 427, 2015
|
|
2LR0
 
 | | Solution NMR structure of de novo designed protein, p-loop ntpase fold, northeast structural genomics consortium target or136 | | Descriptor: | P-loop ntpase fold | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Lee, H, Janjua, H, Kohan, E, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-03-19 | | Release date: | 2012-07-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of de novo designed protein, p-loop ntpase fold, northeast structural genomics consortium target or136
To be Published
|
|
2LMD
 
 | | Minimal Constraints Solution NMR Structure of Prospero Homeobox protein 1 from Homo sapiens, Northeast Structural Genomics Consortium Target HR4660B | | Descriptor: | Prospero homeobox protein 1 | | Authors: | Rossi, P, Lange, O.A, Lee, H, Maglaqui, M, Janjua, H, Ciccosanti, C, Zhao, L, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-11-29 | | Release date: | 2011-12-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5GV5
 
 | | Crystal structure of Candida antarctica Lipase B with active Ser105 modified with a phosphonate inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase B, ... | | Authors: | Park, S.Y, Lee, H. | | Deposit date: | 2016-09-02 | | Release date: | 2017-09-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural and Experimental Evidence for the Enantiomeric Recognition toward a Bulky sec-Alcohol by Candida antarctica Lipase B
Acs Catalysis, 6, 2016
|
|
