3S44
 
 | | Crystal Structure of Pasteurella multocida sialyltransferase M144D mutant with CMP bound | | 分子名称: | Alpha-2,3/2,6-sialyltransferase/sialidase, CMP-3F(a)-Neu5Ac | | 著者 | Sugiarto, G, Lau, K, Li, Y, Lim, S, Ames, J.B, Le, D.-T, Fisher, A.J, Chen, X. | | 登録日 | 2011-05-18 | | 公開日 | 2012-08-15 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | A Sialyltransferase Mutant with Decreased Donor Hydrolysis and Reduced Sialidase Activities for Directly Sialylating Lewis(x).
Acs Chem.Biol., 7, 2012
|
|
7QUV
 
 | | Crystal structure of human Calprotectin (S100A8/S100A9) in complex with Peptide 3 | | 分子名称: | 1,2-ETHANEDIOL, 4-methanoyl-2-(6-oxidanyl-3-oxidanylidene-4~{H}-xanthen-9-yl)benzoic acid, AMINO GROUP, ... | | 著者 | Diaz-Perlas, C, Heinis, C, Pojer, F, Lau, K. | | 登録日 | 2022-01-19 | | 公開日 | 2023-02-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | High-affinity peptides developed against calprotectin and their application as synthetic ligands in diagnostic assays.
Nat Commun, 14, 2023
|
|
8PSD
 
 | | SARS-CoV-2 XBB 1.0 closed conformation. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Duhoo, Y, Lau, K. | | 登録日 | 2023-07-13 | | 公開日 | 2023-11-01 | | 最終更新日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Broadly potent anti-SARS-CoV-2 antibody shares 93% of epitope with ACE2 and provides full protection in monkeys.
J Infect, 87, 2023
|
|
7ZAK
 
 | | Crystal structure of HLA-DP (DPA1*02:01-DPB1*01:01) in complex with a peptide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Racle, J, Guillaume, P, Larabi, A, Lau, K, Pojer, F, Gfeller, D. | | 登録日 | 2022-03-22 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Machine learning predictions of MHC-II specificities reveal alternative binding mode of class II epitopes.
Immunity, 56, 2023
|
|
8PQ2
 
 | | XBB 1.0 RBD bound to P4J15 (Local) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, P4J15 Fragment Antigen-Binding Heavy Chain, P4J15 Fragment Antigen-Binding Light Chain, ... | | 著者 | Duhoo, Y, Lau, K. | | 登録日 | 2023-07-10 | | 公開日 | 2023-11-01 | | 最終更新日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (3.85 Å) | | 主引用文献 | Broadly potent anti-SARS-CoV-2 antibody shares 93% of epitope with ACE2 and provides full protection in monkeys.
J Infect, 87, 2023
|
|
4ERT
 
 | |
7ZFR
 
 | | Crystal structure of HLA-DP (DPA1*02:01-DPB1*01:01) in complex with a peptide bound in the reverse direction | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MHC class II HLA-DP alpha chain (DPA1*02:01), MHC class II HLA-DP beta chain (DPB1*01:01), ... | | 著者 | Racle, J, Guillaume, P, Larabi, A, Lau, K, Pojer, F, Gfeller, D. | | 登録日 | 2022-04-01 | | 公開日 | 2023-04-12 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Machine learning predictions of MHC-II specificities reveal alternative binding mode of class II epitopes.
Immunity, 56, 2023
|
|
4ETU
 
 | |
4ETT
 
 | |
4ETV
 
 | |
4ESU
 
 | |
4ERV
 
 | |
6QTS
 
 | |
6QTR
 
 | |
6QTT
 
 | |
6QTQ
 
 | |
6QTX
 
 | |
6QTV
 
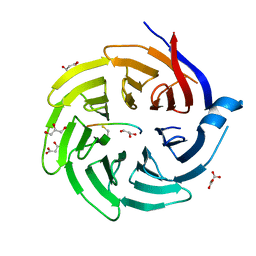 | |
6QTO
 
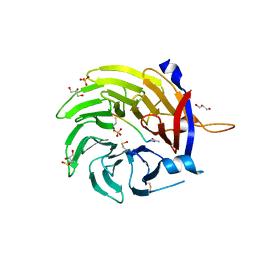 | |
6QTW
 
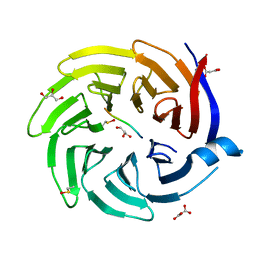 | |
6QTU
 
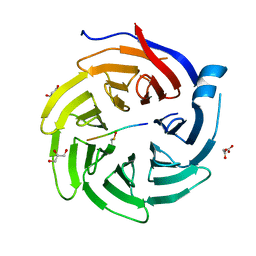 | |
3P9U
 
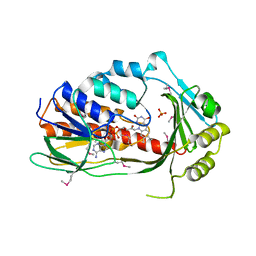 | | Crystal structure of TetX2 from Bacteroides thetaiotaomicron with substrate analogue | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, TetX2 protein | | 著者 | Walkiewicz, K, Davlieva, M, Sun, C, Lau, K, Shamoo, Y. | | 登録日 | 2010-10-18 | | 公開日 | 2011-04-27 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Crystal structure of Bacteroides thetaiotaomicron TetX2: a tetracycline degrading monooxygenase at 2.8 A resolution.
Proteins, 79, 2011
|
|
7ZBB
 
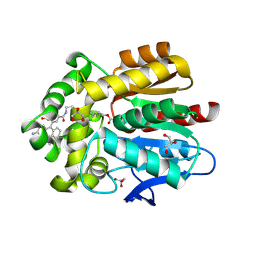 | | HaloTag with TRaQ-G-ctrl ligand | | 分子名称: | (E)-[7-azanyl-10-[2-carboxy-5-[2-[2-(6-chloranylhexoxy)ethoxy]ethylcarbamoyl]phenyl]-5,5-dimethyl-benzo[b][1]benzosilin-3-ylidene]-methyl-azanium, CHLORIDE ION, GLYCEROL, ... | | 著者 | Emmert, S, Rivera-Fuentes, P, Pojer, F, Lau, K. | | 登録日 | 2022-03-23 | | 公開日 | 2023-02-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | A locally activatable sensor for robust quantification of organellar glutathione.
Nat.Chem., 15, 2023
|
|
7ZBA
 
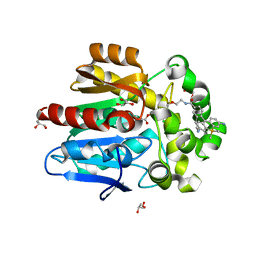 | | HaloTag with Me-TRaQ-G ligand | | 分子名称: | 4-(7-azanyl-5,5-dimethyl-3-methylimino-benzo[b][1]benzosilin-10-yl)-N-[2-[2-(6-chloranylhexoxy)ethoxy]ethyl]-3-methyl-benzamide, CHLORIDE ION, GLYCEROL, ... | | 著者 | Emmert, S, Rivera-Fuentes, P, Pojer, F, Lau, K. | | 登録日 | 2022-03-23 | | 公開日 | 2023-02-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.23 Å) | | 主引用文献 | A locally activatable sensor for robust quantification of organellar glutathione.
Nat.Chem., 15, 2023
|
|
7ZBD
 
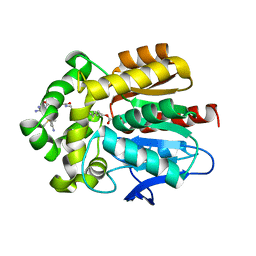 | | HaloTag with TRaQ-G ligand | | 分子名称: | (10R)-7-azanyl-N-[2-[2-(6-chloranylhexoxy)ethoxy]ethyl]-2'-cyano-5,5-dimethyl-3-(methylamino)-1'-oxidanylidene-spiro[benzo[b][1]benzosiline-10,3'-isoindole]-5'-carboxamide, CHLORIDE ION, GLYCEROL, ... | | 著者 | Emmert, S, Rivera-Fuentes, P, Pojer, F, Lau, K. | | 登録日 | 2022-03-23 | | 公開日 | 2023-02-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | A locally activatable sensor for robust quantification of organellar glutathione.
Nat.Chem., 15, 2023
|
|
