6UZB
 
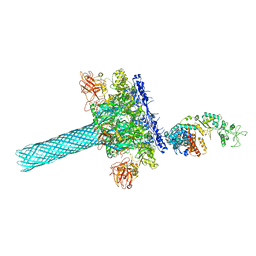 | | Anthrax toxin protective antigen channels bound to edema factor | | Descriptor: | CALCIUM ION, Calmodulin-sensitive adenylate cyclase, Protective antigen | | Authors: | Hardenbrook, N.J, Liu, S, Zhou, K, Zhou, Z.H, Krantz, B.A. | | Deposit date: | 2019-11-14 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Atomic structures of anthrax toxin protective antigen channels bound to partially unfolded lethal and edema factors.
Nat Commun, 11, 2020
|
|
4HQ1
 
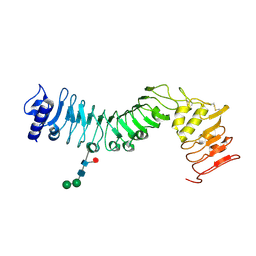 | | Crystal structure of an LRR protein with two solenoids | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Probable receptor protein kinase TMK1, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chai, J, Liu, P, Hu, Z, Zhou, B, Liu, S. | | Deposit date: | 2012-10-25 | | Release date: | 2013-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of an LRR protein with two solenoids
Cell Res., 23, 2013
|
|
6UZE
 
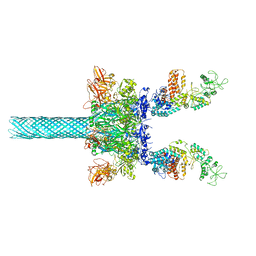 | | Anthrax toxin protective antigen channels bound to edema factor | | Descriptor: | CALCIUM ION, Calmodulin-sensitive adenylate cyclase, Protective antigen | | Authors: | Hardenbrook, N.J, Liu, S, Zhou, K, Zhou, Z.H, Krantz, B.A. | | Deposit date: | 2019-11-15 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Atomic structures of anthrax toxin protective antigen channels bound to partially unfolded lethal and edema factors.
Nat Commun, 11, 2020
|
|
6UZD
 
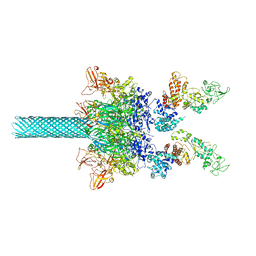 | | Anthrax toxin protective antigen channels bound to edema factor | | Descriptor: | CALCIUM ION, Calmodulin-sensitive adenylate cyclase, Protective antigen | | Authors: | Hardenbrook, N.J, Liu, S, Zhou, K, Zhou, Z.H, Krantz, B.A. | | Deposit date: | 2019-11-14 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Atomic structures of anthrax toxin protective antigen channels bound to partially unfolded lethal and edema factors.
Nat Commun, 11, 2020
|
|
5JJU
 
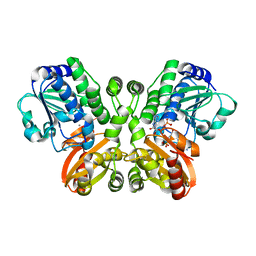 | | Crystal structure of Rv2837c complexed with 5'-pApA and 5'-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, RNA (5'-R(P*AP*A)-3'), ... | | Authors: | Wang, F, He, Q, Liu, S, Gu, L. | | Deposit date: | 2016-04-25 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.312 Å) | | Cite: | Structural and biochemical insight into the mechanism of Rv2837c from Mycobacterium tuberculosis as a c-di-NMP phosphodiesterase
J.Biol.Chem., 291, 2016
|
|
6VRA
 
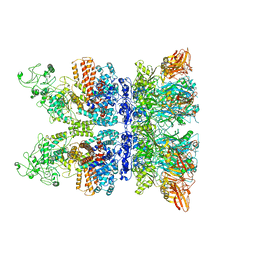 | | Anthrax octamer prechannel bound to full-length edema factor | | Descriptor: | CALCIUM ION, Calmodulin-sensitive adenylate cyclase, Protective antigen | | Authors: | Zhou, K, Hardenbrook, N.J, Liu, S, Cui, Y.X, Krantz, B.A, Zhou, Z.H. | | Deposit date: | 2020-02-07 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic Structures of Anthrax Prechannel Bound with Full-Length Lethal and Edema Factors.
Structure, 28, 2020
|
|
3TEF
 
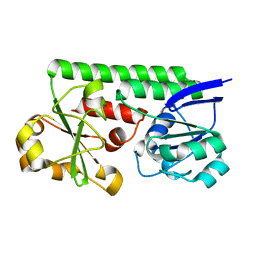 | | Crystal Structure of the Periplasmic Catecholate-Siderophore Binding Protein VctP from Vibrio Cholerae | | Descriptor: | Iron(III) ABC transporter, periplasmic iron-compound-binding protein | | Authors: | Liu, X, Wang, Z, Liu, S, Li, N, Chen, Y, Zhu, C, Zhu, D, Wei, T, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-08-13 | | Release date: | 2012-08-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Crystal structure of periplasmic catecholate-siderophore binding protein VctP from Vibrio cholerae at 1.7 A resolution
Febs Lett., 586, 2012
|
|
5JWV
 
 | | T4 Lysozyme L99A/M102Q with Ethylbenzene Bound | | Descriptor: | Endolysin, PHENYLETHANE | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
1RFA
 
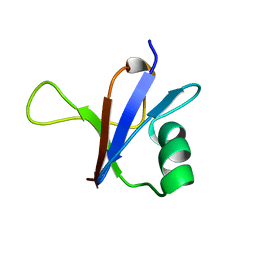 | | NMR SOLUTION STRUCTURE OF THE RAS-BINDING DOMAIN OF C-RAF-1 | | Descriptor: | RAF1 | | Authors: | Emerson, S.D, Madison, V.S, Palermo, R.E, Waugh, D.S, Scheffler, J.E, Tsao, K.-L, Kiefer, S.E, Liu, S.P, Fry, D.C. | | Deposit date: | 1995-04-26 | | Release date: | 1996-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Ras-binding domain of c-Raf-1 and identification of its Ras interaction surface.
Biochemistry, 34, 1995
|
|
5JWT
 
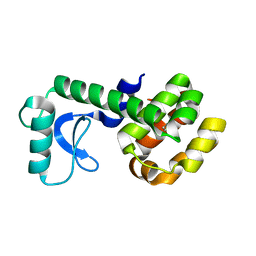 | | T4 Lysozyme L99A/M102Q with Benzene Bound | | Descriptor: | BENZENE, Endolysin | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
5JWU
 
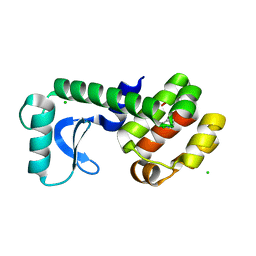 | | T4 Lysozyme L99A/M102Q with 1,2-Dihydro-1,2-azaborine Bound | | Descriptor: | 1,2-dihydro-1,2-azaborinine, CHLORIDE ION, Endolysin | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
2BKU
 
 | | Kap95p:RanGTP complex | | Descriptor: | GTP-BINDING NUCLEAR PROTEIN RAN, GUANOSINE-5'-TRIPHOSPHATE, IMPORTIN BETA-1 SUBUNIT, ... | | Authors: | Lee, S.J, Matsuura, Y, Liu, S.M, Stewart, M. | | Deposit date: | 2005-02-21 | | Release date: | 2005-05-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Nuclear Import Complex Dissociation by Rangtp
Nature, 435, 2005
|
|
8OW8
 
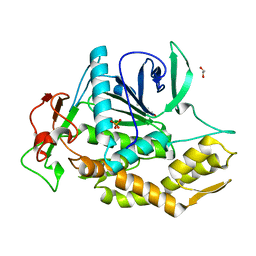 | | Crystal Structure of the Catalytic Domain of a Botulinum Neurotoxin Homologue from Enterococcus faecium | | Descriptor: | 1,2-ETHANEDIOL, Botulinum-like toxin eBoNT/J light chain, PHOSPHATE ION, ... | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M. | | Deposit date: | 2023-04-27 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Catalytic Domain of a Botulinum Neurotoxin Homologue from Enterococcus faecium : Potential Insights into Substrate Recognition.
Int J Mol Sci, 24, 2023
|
|
4DGP
 
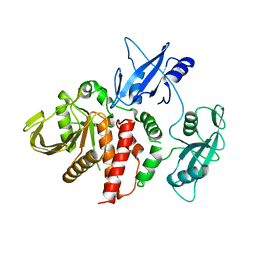 | | The wild-type Src homology 2 (SH2)-domain containing protein tyrosine phosphatase-2 (SHP2) | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Yu, Z.H, Xu, J, Walls, C.D, Chen, L, Zhang, S, Wu, L, Wang, L.N, Liu, S.J, Zhang, Z.Y. | | Deposit date: | 2012-01-26 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Mechanistic Insights into LEOPARD Syndrome-Associated SHP2 Mutations.
J.Biol.Chem., 288, 2013
|
|
4DGX
 
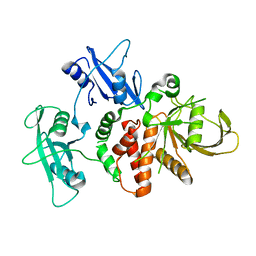 | | LEOPARD Syndrome-Associated SHP2/Y279C mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Yu, Z.H, Xu, J, Walls, C.D, Chen, L, Zhang, S, Wu, L, Wang, L.N, Liu, S.J, Zhang, Z.Y. | | Deposit date: | 2012-01-27 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Mechanistic Insights into LEOPARD Syndrome-Associated SHP2 Mutations.
J.Biol.Chem., 288, 2013
|
|
7TW5
 
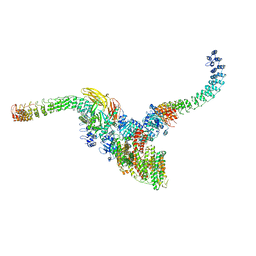 | | Cryo-EM structure of human ankyrin complex (B2P1A2) from red blood cell | | Descriptor: | Ankyrin-1, Band 3 anion transport protein, Protein 4.2 | | Authors: | Xia, X, Liu, S.H, Zhou, Z.H. | | Deposit date: | 2022-02-06 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structure, dynamics and assembly of the ankyrin complex on human red blood cell membrane.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TW6
 
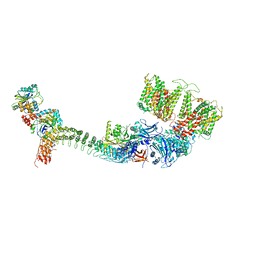 | | Cryo-EM structure of human ankyrin complex (B4P1A1) from red blood cell | | Descriptor: | Ankyrin-1, Band 3 anion transport protein, Protein 4.2 | | Authors: | Xia, X, Liu, S.H, Zhou, Z.H. | | Deposit date: | 2022-02-06 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structure, dynamics and assembly of the ankyrin complex on human red blood cell membrane.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TVZ
 
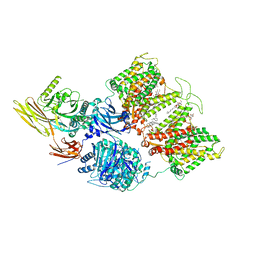 | | Cryo-EM structure of human band 3-protein 4.2 complex in diagonal conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, CHOLESTEROL, ... | | Authors: | Xia, X, Liu, S.H, Zhou, Z.H. | | Deposit date: | 2022-02-06 | | Release date: | 2022-06-08 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure, dynamics and assembly of the ankyrin complex on human red blood cell membrane.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TW3
 
 | | Cryo-EM structure of human ankyrin complex (B2P1A1) from red blood cell | | Descriptor: | Ankyrin-1, Band 3 anion transport protein, Protein 4.2 | | Authors: | Xia, X, Liu, S.H, Zhou, Z.H. | | Deposit date: | 2022-02-06 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure, dynamics and assembly of the ankyrin complex on human red blood cell membrane.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TW2
 
 | |
7TW1
 
 | |
7TW0
 
 | | Cryo-EM structure of human band 3-protein 4.2 complex in vertical conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, Protein 4.2 | | Authors: | Xia, X, Liu, S.H, Zhou, Z.H. | | Deposit date: | 2022-02-06 | | Release date: | 2022-06-08 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure, dynamics and assembly of the ankyrin complex on human red blood cell membrane.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5UPH
 
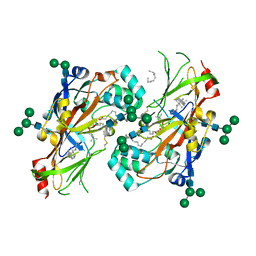 | | Lipids bound lysosomal integral membrane protein 2 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-phosphono-beta-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Conrad, K.S, Liu, S. | | Deposit date: | 2017-02-03 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Lysosomal integral membrane protein-2 as a phospholipid receptor revealed by biophysical and cellular studies.
Nat Commun, 8, 2017
|
|
5HH9
 
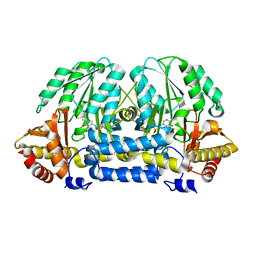 | | Structure of PvdN from Pseudomonas aeruginosa | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, PvdN | | Authors: | Xia, H, Bai, G, Liu, S, Li, N, Xu, S, Chen, K, Yao, Q, Gu, L. | | Deposit date: | 2016-01-10 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural and Biochemical Characterization of a Novel L-Cystine Desulfurase involved in the biosynthesis of the major siderophore pyoverdine in Pseudomonas aeruginosa
To Be Published
|
|
7SLQ
 
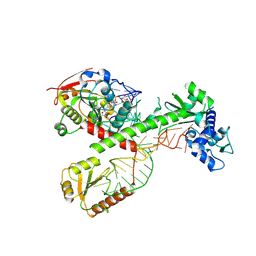 | | Cryo-EM structure of 7SK core RNP with circular RNA | | Descriptor: | 7SK snRNA methylphosphate capping enzyme, La-related protein 7, Minimal circular 7SK RNA, ... | | Authors: | Yang, Y, Liu, S, Zhou, Z.H, Feigon, J. | | Deposit date: | 2021-10-24 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of RNA conformational switching in the transcriptional regulator 7SK RNP.
Mol.Cell, 82, 2022
|
|
