8AGK
 
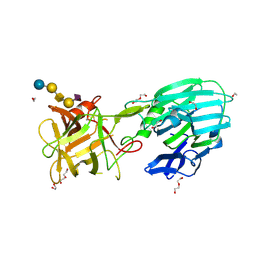 | | Botulinum neurotoxin subtype A6 cell binding domain in complex with GD1a ganglioside | | 分子名称: | 1,2-ETHANEDIOL, Bont/A1, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Gregory, K.S, Acharya, K.R, Liu, S.M, Newell, A.R, Mojanaga, O.O. | | 登録日 | 2022-07-20 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal Structures of the Clostridium botulinum Neurotoxin A6 Cell Binding Domain Alone and in Complex with GD1a Reveal Significant Conformational Flexibility.
Int J Mol Sci, 23, 2022
|
|
8ALP
 
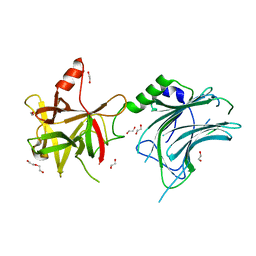 | | Botulinum neurotoxin A6 cell binding domain crystal form II | | 分子名称: | 1,2-ETHANEDIOL, Bont/A1, DI(HYDROXYETHYL)ETHER | | 著者 | Gregory, K.S, Acharya, K.R, Liu, S.M. | | 登録日 | 2022-08-01 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal Structures of the Clostridium botulinum Neurotoxin A6 Cell Binding Domain Alone and in Complex with GD1a Reveal Significant Conformational Flexibility.
Int J Mol Sci, 23, 2022
|
|
4B4A
 
 | | Structure of the TatC core of the twin arginine protein translocation system | | 分子名称: | Lauryl Maltose Neopentyl Glycol, SEC-INDEPENDENT PROTEIN TRANSLOCASE PROTEIN TATC | | 著者 | Rollauer, S.E, Tarry, M.J, Jaaskelainen, M, Graham, J.E, Jaeger, F, Krehenbrink, M, Roversi, P, McDowell, M.A, Stansfeld, P.J, Johnson, S, Liu, S.M, Lukey, M.J, Marcoux, J, Robinson, C.V, Sansom, M.S, Palmer, T, Hogbom, M, Berks, B.C, Lea, S.M. | | 登録日 | 2012-07-30 | | 公開日 | 2012-12-05 | | 最終更新日 | 2012-12-19 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structure of the Tatc Core of the Twin-Arginine Protein Transport System.
Nature, 49, 2012
|
|
4ZMU
 
 | | Dcsbis, a diguanylate cyclase from Pseudomonas aeruginosa | | 分子名称: | diguanylate cyclase | | 著者 | Chen, Y, Liu, C, Liu, S, Chi, K, Gu, L. | | 登録日 | 2015-05-04 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.502 Å) | | 主引用文献 | crystal structure of Dcsbis from Pseudomonas aeruginosa
To Be Published
|
|
4ZMM
 
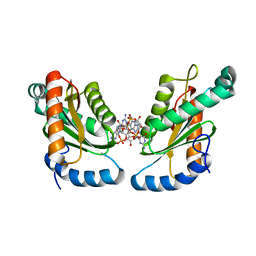 | | GGDEF domain of Dcsbis complexed with c-di-GMP | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), diguanylate cyclase | | 著者 | Chen, Y, Liu, C, Liu, S, Chi, K, Gu, L. | | 登録日 | 2015-05-04 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.503 Å) | | 主引用文献 | Crystal structure of Dcsbis GGDEF domain complexed with c-di-GMP
To Be Published
|
|
6G5G
 
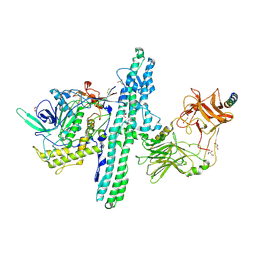 | | Crystal structure of an engineered Botulinum Neurotoxin type B mutant E1191M/S1199Y in complex with human synaptotagmin 2 | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | 著者 | Masuyer, G, Elliot, M, Favre-Guilmard, C, Liu, S.M, Maignel, J, Beard, M, Carre, D, Kalinichev, M, Lezmi, S, Mir, I, Nicoleau, C, Palan, S, Perier, C, Raban, E, Dong, M, Krupp, J, Stenmark, P. | | 登録日 | 2018-03-29 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Engineered botulinum neurotoxin B with improved binding to human receptors has enhanced efficacy in preclinical models.
Sci Adv, 5, 2019
|
|
6G5F
 
 | | Crystal structure of an engineered Botulinum Neurotoxin type B mutant E1191M/S1199Y in complex with human synaptotagmin 1 | | 分子名称: | Botulinum neurotoxin type B, GLYCEROL, MALONATE ION, ... | | 著者 | Masuyer, G, Elliot, M, Favre-Guilmard, C, Liu, S.M, Maignel, J, Beard, M, Carre, D, Kalinichev, M, Lezmi, S, Mir, I, Nicoleau, C, Palan, S, Perier, C, Raban, E, Dong, M, Krupp, J, Stenmark, P. | | 登録日 | 2018-03-29 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Engineered botulinum neurotoxin B with improved binding to human receptors has enhanced efficacy in preclinical models.
Sci Adv, 5, 2019
|
|
4WXW
 
 | |
6G5K
 
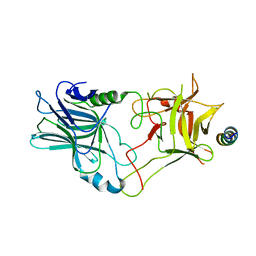 | | Crystal structure of the binding domain of Botulinum Neurotoxin type B in complex with human synaptotagmin 1 | | 分子名称: | Botulinum neurotoxin type B, Synaptotagmin-1 | | 著者 | Masuyer, G, Elliot, M, Favre-Guilmard, C, Liu, S.M, Maignel, J, Beard, M, Carre, D, Kalinichev, M, Lezmi, S, Mir, I, Nicoleau, C, Palan, S, Perier, C, Raban, E, Dong, M, Krupp, J, Stenmark, P. | | 登録日 | 2018-03-29 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Engineered botulinum neurotoxin B with improved binding to human receptors has enhanced efficacy in preclinical models.
Sci Adv, 5, 2019
|
|
4WXO
 
 | |
5JWT
 
 | | T4 Lysozyme L99A/M102Q with Benzene Bound | | 分子名称: | BENZENE, Endolysin | | 著者 | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | 登録日 | 2016-05-12 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
5GT2
 
 | | Crystal Structure and Biochemical Features of dye-decolorizing peroxidase YfeX from Escherichia coli O157 | | 分子名称: | PROTOPORPHYRIN IX CONTAINING FE, Probable deferrochelatase/peroxidase YfeX | | 著者 | Ma, Y.L, Yuan, Z.G, Liu, S, Wang, J.X, Gu, L.C, Liu, X.H. | | 登録日 | 2016-08-18 | | 公開日 | 2017-02-08 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.093 Å) | | 主引用文献 | Crystal structure and biochemical features of dye-decolorizing peroxidase YfeX from Escherichia coli O157 Asp(143) and Arg(232) play divergent roles toward different substrates
Biochem. Biophys. Res. Commun., 484, 2017
|
|
5JWU
 
 | | T4 Lysozyme L99A/M102Q with 1,2-Dihydro-1,2-azaborine Bound | | 分子名称: | 1,2-dihydro-1,2-azaborinine, CHLORIDE ION, Endolysin | | 著者 | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | 登録日 | 2016-05-12 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
1MRH
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | 分子名称: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, ALPHA-MOMORCHARIN | | 著者 | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | 登録日 | 1994-07-01 | | 公開日 | 1995-02-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1MRK
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | 分子名称: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, ALPHA-TRICHOSANTHIN | | 著者 | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | 登録日 | 1994-07-01 | | 公開日 | 1995-02-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
5JWS
 
 | | T4 Lysozyme L99A with 1-Hydro-2-ethyl-1,2-azaborine Bound | | 分子名称: | 2-ethyl-1,2-dihydro-1,2-azaborinine, CHLORIDE ION, Endolysin | | 著者 | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | 登録日 | 2016-05-12 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
1MRI
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | 分子名称: | ALPHA-MOMORCHARIN | | 著者 | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | 登録日 | 1994-07-01 | | 公開日 | 1995-02-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
5JWW
 
 | | T4 Lysozyme L99A/M102Q with 1-Hydro-2-ethyl-1,2-azaborine Bound | | 分子名称: | 2-ethyl-1,2-dihydro-1,2-azaborinine, CHLORIDE ION, Endolysin | | 著者 | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | 登録日 | 2016-05-12 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
6WNL
 
 | | human Artemis/SNM1C catalytic domain, crystal form 2 | | 分子名称: | Protein artemis, ZINC ION | | 著者 | Karim, F, Liu, S, Laciak, A.R, Volk, L, Rosenblum, M, Curtis, R, Huang, N, Carr, G, Zhu, G. | | 登録日 | 2020-04-22 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Structural analysis of the catalytic domain of Artemis endonuclease/SNM1C reveals distinct structural features.
J.Biol.Chem., 295, 2020
|
|
6WO0
 
 | | human Artemis/SNM1C catalytic domain, crystal form 1 | | 分子名称: | GLYCEROL, Protein artemis, ZINC ION | | 著者 | Karim, F, Liu, S, Laciak, A.R, Volk, L, Rosenblum, M, Curtis, R, Huang, N, Carr, G, Zhu, G. | | 登録日 | 2020-04-23 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structural analysis of the catalytic domain of Artemis endonuclease/SNM1C reveals distinct structural features.
J.Biol.Chem., 295, 2020
|
|
1MRG
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | 分子名称: | ADENOSINE, ALPHA-MOMORCHARIN | | 著者 | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | 登録日 | 1994-07-01 | | 公開日 | 1995-02-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
5JWV
 
 | | T4 Lysozyme L99A/M102Q with Ethylbenzene Bound | | 分子名称: | Endolysin, PHENYLETHANE | | 著者 | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | 登録日 | 2016-05-12 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
1MRJ
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | 分子名称: | ADENOSINE, ALPHA-TRICHOSANTHIN | | 著者 | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | 登録日 | 1994-07-01 | | 公開日 | 1995-02-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1PJD
 
 | | Structure and Topology of a Peptide Segment of the 6th Transmembrane Domain of the Saccharomyces cerevisiae alpha-Factor Receptor in Phospholipid Bilayers | | 分子名称: | Pheromone alpha factor receptor | | 著者 | Valentine, K.G, Liu, S.-F, Marassi, F.M, Veglia, G, Nevzorov, A.A, Opella, S.J, Ding, F.-X, Wang, S.-H, Arshava, B, Becker, J.M, Naider, F. | | 登録日 | 2003-06-02 | | 公開日 | 2003-09-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Structure and Topology of a Peptide Segment of the 6th Transmembrane Domain of the Saccharomyces cerevisiae alpha-Factor Receptor in Phospholipid Bilayers
Biopolymers, 59, 2001
|
|
5JJU
 
 | | Crystal structure of Rv2837c complexed with 5'-pApA and 5'-AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, RNA (5'-R(P*AP*A)-3'), ... | | 著者 | Wang, F, He, Q, Liu, S, Gu, L. | | 登録日 | 2016-04-25 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.312 Å) | | 主引用文献 | Structural and biochemical insight into the mechanism of Rv2837c from Mycobacterium tuberculosis as a c-di-NMP phosphodiesterase
J.Biol.Chem., 291, 2016
|
|
