8TBX
 
 | | Crystal structure of human DDX1 helicase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DDX1, MAGNESIUM ION, ... | | Authors: | Zeng, H, Dong, A, Li, Y, Yen, H, Hejazi, Z, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Halabelian, L. | | Deposit date: | 2023-06-29 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of human DDX1 helicase in complex with ADP
To be published
|
|
3GF9
 
 | | Crystal structure of human Intersectin 2 RhoGEF domain | | Descriptor: | Intersectin 2, UNKNOWN ATOM OR ION | | Authors: | Shen, Y, Tong, Y, Tempel, W, Li, Y, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-02-26 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human Intersectin 2 RhoGEF domain
To be Published
|
|
8HQT
 
 | |
8HQV
 
 | |
8HQW
 
 | |
7XWX
 
 | | Crystal structure of SARS-CoV-2 N-CTD | | Descriptor: | Nucleoprotein, PHOSPHATE ION | | Authors: | Luan, X.D, Li, X.M, Li, Y.F. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
7XX1
 
 | | Crystal structure of SARS-CoV-2 N-NTD | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nucleoprotein, ZINC ION | | Authors: | Luan, X.D, Li, X.M, Li, Y.F. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
8GCY
 
 | | Co-crystal structure of CBL-B in complex with N-Aryl isoindolin-1-one inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-{3-[(1s,3R)-3-methyl-1-(4-methyl-4H-1,2,4-triazol-3-yl)cyclobutyl]phenyl}-6-{[(3S)-3-methylpiperidin-1-yl]methyl}-4-(trifluoromethyl)-2,3-dihydro-1H-isoindol-1-one, E3 ubiquitin-protein ligase CBL-B, ... | | Authors: | Kimani, S, Zeng, H, Dong, A, Li, Y, Santhakumar, V, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The co-crystal structure of Cbl-b and a small-molecule inhibitor reveals the mechanism of Cbl-b inhibition.
Commun Biol, 6, 2023
|
|
8HR0
 
 | | The complex structure of COPII coat with HCoV-OC43 DD sorting motif | | Descriptor: | HCoV-OC43, Protein transport protein Sec23A, Protein transport protein Sec24A, ... | | Authors: | Ma, W.F, Nan, Y.N, Yang, M.R, Li, Y.Q. | | Deposit date: | 2022-12-14 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Tighter ER retention of SARS-CoV-2 Omicron spike caused by a constellation of folding disruptive mutations
To Be Published
|
|
3CWD
 
 | | Molecular recognition of nitro-fatty acids by PPAR gamma | | Descriptor: | (9E,12Z)-10-nitrooctadeca-9,12-dienoic acid, (9Z,12E)-12-nitrooctadeca-9,12-dienoic acid, Peroxisome proliferator-activated receptor gamma, ... | | Authors: | Martynowski, D, Li, Y. | | Deposit date: | 2008-04-21 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular recognition of nitrated fatty acids by PPAR gamma.
Nat.Struct.Mol.Biol., 15, 2008
|
|
8TRE
 
 | | Crystal structure of the Human TRIP12 WWE domain (isoform 2) in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Isoform 2 of E3 ubiquitin-protein ligase TRIP12, UNKNOWN ATOM OR ION | | Authors: | Kimani, S, Dong, A, Li, Y, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-08-09 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the Human TRIP12 WWE domain (isoform 2) in complex with ATP
To be published
|
|
7MWL
 
 | | The TAM domain of BAZ2A in complex with a 12mer mCG DNA | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), GLYCEROL | | Authors: | Liu, K, Dong, A, Li, Y, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-17 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The TAM domain of BAZ2A in complex with a 12mer mCG DNA
To Be Published
|
|
6PE4
 
 | | Yeast Vo motor in complex with 1 VopQ molecule | | Descriptor: | Cation transporter, Uncharacterized protein YPR170W-B, V-type proton ATPase subunit a, ... | | Authors: | Peng, W, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2019-06-20 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A distinct inhibitory mechanism of the V-ATPase by Vibrio VopQ revealed by cryo-EM.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6IYG
 
 | | The Structure of Maltooligosaccharide-forming Amylase from Pseudomonas saccharophila STB07 with Maltotetraose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glucan 1,4-alpha-maltotetraohydrolase, ... | | Authors: | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | Deposit date: | 2018-12-15 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Maltotetraose-forming amylase from Pseudomonas saccharophila STB07
To Be Published
|
|
8Q72
 
 | | E. coli plasmid-borne JetABCD(E248A) core in a cleavage-competent state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Circular plasmid DNA (1840-MER), JetA, ... | | Authors: | Roisne-Hamelin, F, Li, Y, Gruber, S. | | Deposit date: | 2023-08-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Structural basis for plasmid restriction by SMC JET nuclease.
Mol.Cell, 84, 2024
|
|
7M3X
 
 | | Crystal Structure of the Apo Form of Human RBBP7 | | Descriptor: | Histone-binding protein RBBP7, UNKNOWN ATOM OR ION | | Authors: | Righetto, G.L, Dong, A, Li, Y, Hutchinson, A, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-19 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal Structure of the Apo Form of Human RBBP7
To Be Published
|
|
8TLU
 
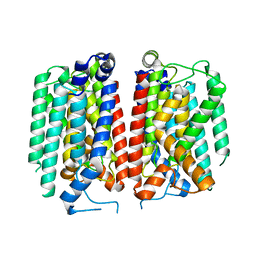 | | E. coli MraY mutant-T23P | | Descriptor: | Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Orta, A.K, Li, Y.E, Clemons, W.M. | | Deposit date: | 2023-07-27 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Synthesis of lipid-linked precursors of the bacterial cell wall is governed by a feedback control mechanism in Pseudomonas aeruginosa.
Nat Microbiol, 9, 2024
|
|
1FC4
 
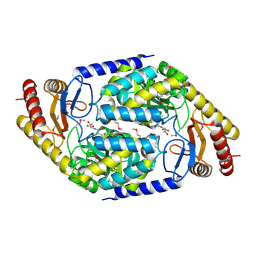 | | 2-AMINO-3-KETOBUTYRATE COA LIGASE | | Descriptor: | 2-AMINO-3-KETOBUTYRATE CONENZYME A LIGASE, 2-AMINO-3-KETOBUTYRIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Schmidt, A, Matte, A, Li, Y, Sivaraman, J, Larocque, R, Schrag, J.D, Smith, C, Sauve, V, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2000-07-17 | | Release date: | 2001-05-02 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of 2-amino-3-ketobutyrate CoA ligase from Escherichia coli complexed with a PLP-substrate intermediate: inferred reaction mechanism.
Biochemistry, 40, 2001
|
|
1G91
 
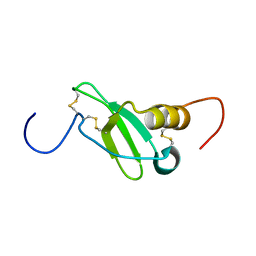 | | SOLUTION STRUCTURE OF MYELOID PROGENITOR INHIBITORY FACTOR-1 (MPIF-1) | | Descriptor: | MYELOID PROGENITOR INHIBITORY FACTOR-1 | | Authors: | Rajarathnam, K, Li, Y, Rohrer, T, Gentz, R. | | Deposit date: | 2000-11-21 | | Release date: | 2001-03-07 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of myeloid progenitor inhibitory factor-1 (MPIF-1), a novel monomeric CC chemokine.
J.Biol.Chem., 276, 2001
|
|
2D34
 
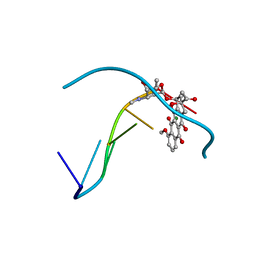 | | FORMALDEHYDE CROSS-LINKS DAUNORUBICIN AND DNA EFFICIENTLY: HPLC AND X-RAY DIFFRACTION STUDIES | | Descriptor: | 5'-D(*CP*GP*TP*(A35)P*CP*G)-3', DAUNOMYCIN, MAGNESIUM ION | | Authors: | Wang, A.H.-J, Gao, Y.-G, Liaw, Y.-C, Li, Y.-K. | | Deposit date: | 1991-05-23 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Formaldehyde cross-links daunorubicin and DNA efficiently: HPLC and X-ray diffraction studies.
Biochemistry, 30, 1991
|
|
3E19
 
 | | Crystal Structure of Iron Uptake Regulatory Protein (FeoA) Solved by Sulfur SAD in a Monoclinic Space Group | | Descriptor: | FeoA, GLYCEROL, PHOSPHATE ION | | Authors: | Hughes, R.C, Li, Y, Wang, B.-C, Liu, Z.-J, Ng, J.D. | | Deposit date: | 2008-08-02 | | Release date: | 2008-12-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Structure Determination of Iron Uptake Regulatory
Protein (FeoA) by Sulfur SAD in a Monoclinic Space Group
To be Published
|
|
263D
 
 | | ISOHELICITY AND PHASING IN DRUG-DNA SEQUENCE RECOGNITION: CRYSTAL STRUCTURE OF A TRIS(BENZIMIDAZOLE)-OLIGONUCLEOTIDE COMPLEX | | Descriptor: | 2''-(4-METHOXYPHENYL)-5-(3-AMINO-1-PYRROLIDINYL)-2,5',2',5''-TRI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Clark, G.R, Gray, E.J, Neidle, S, Li, Y.-H, Leupin, W. | | Deposit date: | 1996-09-27 | | Release date: | 1996-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isohelicity and phasing in drug--DNA sequence recognition: crystal structure of a tris(benzimidazole)--oligonucleotide complex.
Biochemistry, 35, 1996
|
|
2P2O
 
 | | Crystal structure of maltose transacetylase from Geobacillus kaustophilus P2(1) crystal form | | Descriptor: | Maltose transacetylase | | Authors: | Liu, Z.J, Li, Y, Chen, L, Zhu, J, Rose, J.P, Ebihara, A, Yokoyama, S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-07 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of Maltose Transacetylase from Geobacillus Kaustophilus at 1.8 Angstrom Resolution
To be Published
|
|
1Q57
 
 | | The Crystal Structure of the Bifunctional Primase-Helicase of Bacteriophage T7 | | Descriptor: | DNA primase/helicase | | Authors: | Toth, E.A, Li, Y, Sawaya, M.R, Cheng, Y, Ellenberger, T. | | Deposit date: | 2003-08-06 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The Crystal Structure of the Bifunctional Primase-Helicase of Bacteriophage T7
Mol.Cell, 12, 2003
|
|
1JA3
 
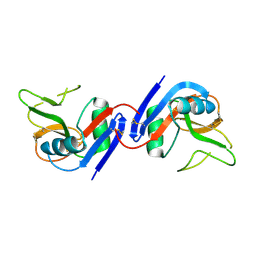 | | Crystal Structure of the Murine NK Cell Inhibitory Receptor Ly-49I | | Descriptor: | MHC class I recognition receptor Ly49I | | Authors: | Dimasi, N, Sawicki, W.M, Reineck, L.A, Li, Y, Natarajan, K, Murgulies, D.H, Mariuzza, A.R. | | Deposit date: | 2001-05-29 | | Release date: | 2002-07-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Ly49I natural killer cell receptor reveals variability in dimerization mode within the Ly49 family.
J.Mol.Biol., 320, 2002
|
|
