5CNY
 
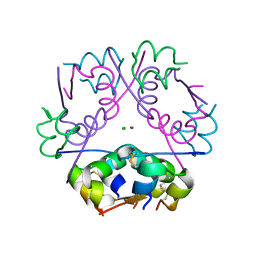 | |
5DCE
 
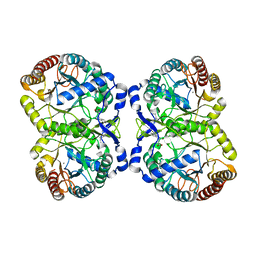 | |
5CZS
 
 | |
5D03
 
 | |
4QH0
 
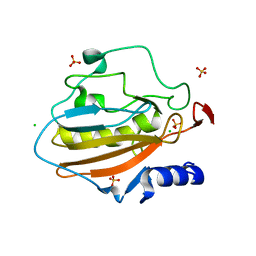 | | Crystal structure of NucA from Streptococcus agalactiae with magnesium ion bound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA-entry nuclease (Competence-specific nuclease), ... | | Authors: | Moon, A.F, Gaudu, P, Pedersen, L.C. | | Deposit date: | 2014-05-26 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of the virulence factor nuclease A from Streptococcus agalactiae.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q5F
 
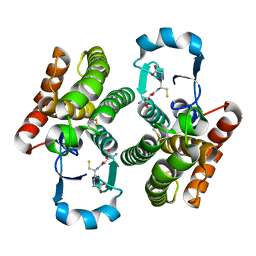 | |
4QGO
 
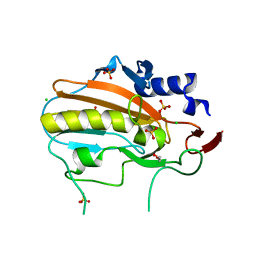 | | Crystal structure of NucA from Streptococcus agalactiae with no metal bound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA-entry nuclease (Competence-specific nuclease), ... | | Authors: | Pedersen, L.C, Moon, A.F, Gaudu, P. | | Deposit date: | 2014-05-23 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural characterization of the virulence factor nuclease A from Streptococcus agalactiae.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4P09
 
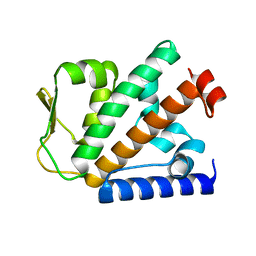 | | Crystal structure of HOIP PUB domain | | Descriptor: | E3 ubiquitin-protein ligase RNF31 | | Authors: | Akutsu, M, Schaeffer, V, Olma, M.H, Gomes, L.C, Kawasaki, M, Dikic, I. | | Deposit date: | 2014-02-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding of OTULIN to the PUB domain of HOIP controls NF-kappa B signaling.
Mol.Cell, 54, 2014
|
|
3FFC
 
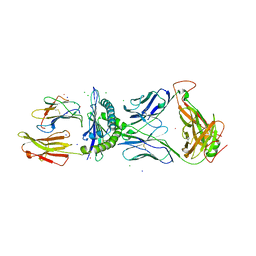 | | Crystal Structure of CF34 TCR in complex with HLA-B8/FLR | | Descriptor: | Beta-2-microglobulin, CADMIUM ION, CF34 alpha chain, ... | | Authors: | Gras, S, Burrows, S.R, Kjer-Nielsen, L, Clements, C.S, Liu, Y.C, Sullivan, L.C, Brooks, A.G, Purcell, A.W, McCluskey, J, Rossjohn, J. | | Deposit date: | 2008-12-03 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The shaping of T cell receptor recognition by self-tolerance.
Immunity, 30, 2009
|
|
1ADN
 
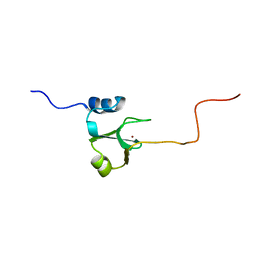 | |
1B71
 
 | | RUBRERYTHRIN | | Descriptor: | FE (III) ION, PROTEIN (RUBRERYTHRIN), ZINC ION | | Authors: | Sieker, L.C, Holmes, M, Le Trong, I, Turley, S, Santarsiero, B.D, Liu, M.-Y, Legall, J, Stenkamp, R.E. | | Deposit date: | 1999-01-26 | | Release date: | 2000-01-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alternative metal-binding sites in rubrerythrin.
Nat.Struct.Biol., 6, 1999
|
|
1BEA
 
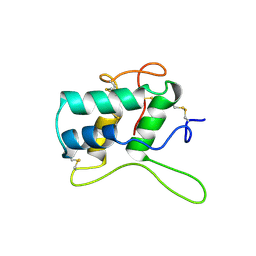 | | BIFUNCTIONAL HAGEMAN FACTOR/AMYLASE INHIBITOR FROM MAIZE | | Descriptor: | BIFUNCTIONAL AMYLASE/SERINE PROTEASE INHIBITOR | | Authors: | Behnke, C.A, Yee, V.C, Le Trong, I, Pedersen, L.C, Stenkamp, R.E, Kim, S.S, Reeck, G.R, Teller, D.C. | | Deposit date: | 1998-05-13 | | Release date: | 1998-08-12 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural determinants of the bifunctional corn Hageman factor inhibitor: x-ray crystal structure at 1.95 A resolution.
Biochemistry, 37, 1998
|
|
1AQU
 
 | |
1TP3
 
 | | PDZ3 domain of PSD-95 protein complexed with KKETPV peptide ligand | | Descriptor: | KKETPV peptide ligand, Presynaptic density protein 95 | | Authors: | Saro, D, Martin, P, Vickrey, J.F, Griffin, A, Kovari, L.C, Spaller, M.R. | | Deposit date: | 2004-06-15 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of the third PDZ domain of PSD-95 protein complexed with KKETPV peptide ligand
To be Published
|
|
3F5F
 
 | | Crystal structure of heparan sulfate 2-O-sulfotransferase from gallus gallus as a maltose binding protein fusion. | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Maltose-binding periplasmic protein, Heparan sulfate 2-O-sulfotransferase 1, ... | | Authors: | Bethea, H.N, Xu, D, Liu, J, Pedersen, L.C. | | Deposit date: | 2008-11-03 | | Release date: | 2008-12-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Redirecting the substrate specificity of heparan sulfate 2-O-sulfotransferase by structurally guided mutagenesis.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3GRB
 
 | | Crystal structure of the F87M/L110M mutant of human transthyretin at pH 6.5 | | Descriptor: | ACETATE ION, GLYCEROL, Transthyretin, ... | | Authors: | Palmieri, L.C, Freire, J.B.B, Foguel, D, Lima, L.M.T.R. | | Deposit date: | 2009-03-25 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Novel Zn2+-binding sites in human transthyretin: implications for amyloidogenesis and retinol-binding protein recognition.
J.Biol.Chem., 285, 2010
|
|
1LVF
 
 | | syntaxin 6 | | Descriptor: | syntaxin 6 | | Authors: | Misura, K.M.S, Bock, J.B, Gonzalez, L.C, Scheller, R.H, Weis, W.I. | | Deposit date: | 2002-05-28 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of the amino-terminal domain of syntaxin 6, a SNAP-25 C homolog.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1UBV
 
 | |
1BFA
 
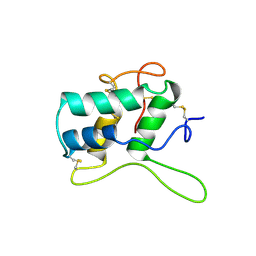 | | RECOMBINANT BIFUNCTIONAL HAGEMAN FACTOR/AMYLASE INHIBITOR FROM MAIZE | | Descriptor: | BIFUNCTIONAL AMYLASE/SERINE PROTEASE INHIBITOR | | Authors: | Behnke, C.A, Yee, V.C, Le Trong, I, Pedersen, L.C, Stenkamp, R.E, Kim, S.S, Reeck, G.R, Teller, D.C. | | Deposit date: | 1998-05-13 | | Release date: | 1998-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural determinants of the bifunctional corn Hageman factor inhibitor: x-ray crystal structure at 1.95 A resolution.
Biochemistry, 37, 1998
|
|
1UBW
 
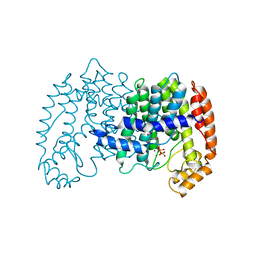 | | STRUCTURE OF FARNESYL PYROPHOSPHATE SYNTHETASE | | Descriptor: | FARNESYL DIPHOSPHATE SYNTHASE, GERANYL DIPHOSPHATE, MAGNESIUM ION | | Authors: | Tarshis, L.C, Proteau, P, Poulter, C.D, Sacchettini, J.C. | | Deposit date: | 1996-10-14 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulation of product chain length by isoprenyl diphosphate synthases.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1UBX
 
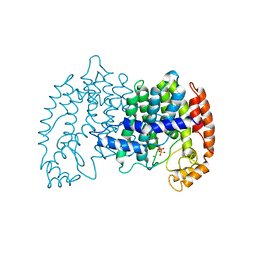 | | STRUCTURE OF FARNESYL PYROPHOSPHATE SYNTHETASE | | Descriptor: | FARNESYL DIPHOSPHATE, FARNESYL DIPHOSPHATE SYNTHASE, MAGNESIUM ION | | Authors: | Tarshis, L.C, Proteau, P, Poulter, C.D, Sacchettini, J.C. | | Deposit date: | 1996-10-14 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulation of product chain length by isoprenyl diphosphate synthases.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1UBY
 
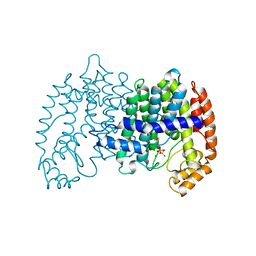 | | STRUCTURE OF FARNESYL PYROPHOSPHATE SYNTHETASE | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, FARNESYL DIPHOSPHATE SYNTHASE, MAGNESIUM ION | | Authors: | Tarshis, L.C, Proteau, P, Poulter, C.D, Sacchettini, J.C. | | Deposit date: | 1996-10-14 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulation of product chain length by isoprenyl diphosphate synthases.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
3H4L
 
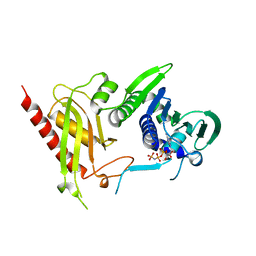 | | Crystal Structure of N terminal domain of a DNA repair protein | | Descriptor: | DNA mismatch repair protein PMS1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Arana, M.E, Holmes, S.F, Fortune, J.M, Moon, A.F, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2009-04-20 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional residues on the surface of the N-terminal domain of yeast Pms1.
Dna Repair, 9, 2010
|
|
1M5A
 
 | |
3H4Z
 
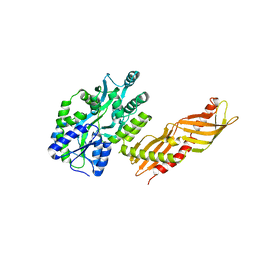 | | Crystal Structure of an MBP-Der p 7 fusion protein | | Descriptor: | Maltose-binding periplasmic protein fused with Allergen DERP7, SODIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Pedersen, L.C, Mueller, G.A, London, R.E. | | Deposit date: | 2009-04-21 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of the dust mite allergen Der p 7 reveals similarities to innate immune proteins.
J.Allergy Clin.Immunol., 125, 2010
|
|
