4AB0
 
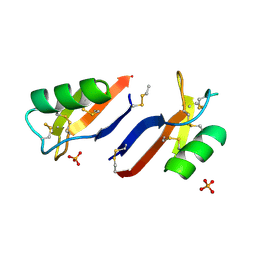 | | X-ray crystal structure of Nicotiana alata defensin NaD1 | | Descriptor: | FLOWER-SPECIFIC DEFENSIN, PHOSPHATE ION | | Authors: | Mills, G.D, Lay, F.T, Hulett, M.D, Kvansakul, M. | | Deposit date: | 2011-12-06 | | Release date: | 2012-04-25 | | Last modified: | 2012-06-20 | | Method: | X-RAY DIFFRACTION (1.636 Å) | | Cite: | Dimerization of Plant Defensin Nad1 Enhances its Antifungal Activity.
J.Biol.Chem., 287, 2012
|
|
5WOU
 
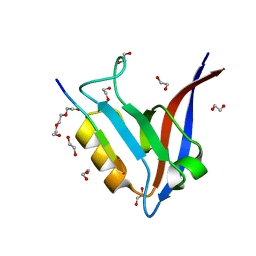 | |
4NXT
 
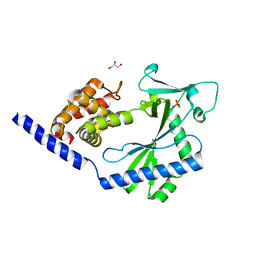 | | Crystal structure of the cytosolic domain of human MiD51 | | Descriptor: | GLYCEROL, Mitochondrial dynamic protein MID51, SULFATE ION | | Authors: | Richter, V, Ryan, M.T, Kvansakul, M. | | Deposit date: | 2013-12-09 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural and functional analysis of MiD51, a dynamin receptor required for mitochondrial fission.
J.Cell Biol., 204, 2014
|
|
4NXV
 
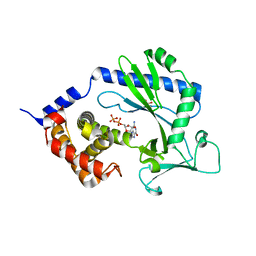 | | Crystal structure of the cytosolic domain of human MiD51 | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, Mitochondrial dynamic protein MID51, ... | | Authors: | Richter, V, Kvansakul, M, Ryan, M.T. | | Deposit date: | 2013-12-09 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of MiD51, a dynamin receptor required for mitochondrial fission.
J.Cell Biol., 204, 2014
|
|
4NXU
 
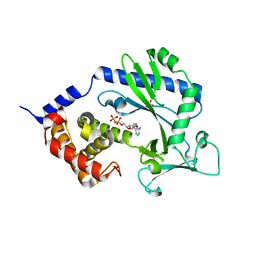 | | Crystal structure of the cytosolic domain of human MiD51 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Mitochondrial dynamic protein MID51, ... | | Authors: | Richter, V, Kvansakul, M, Ryan, M.T. | | Deposit date: | 2013-12-09 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of MiD51, a dynamin receptor required for mitochondrial fission.
J.Cell Biol., 204, 2014
|
|
4NXX
 
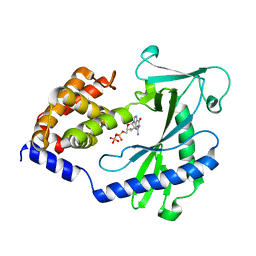 | |
4NXW
 
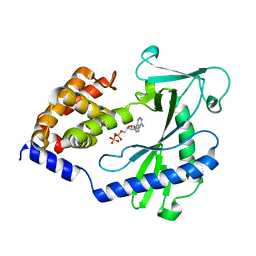 | |
6XA8
 
 | |
6XA6
 
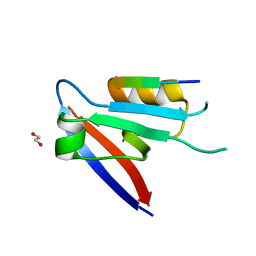 | |
6XA7
 
 | |
6YLI
 
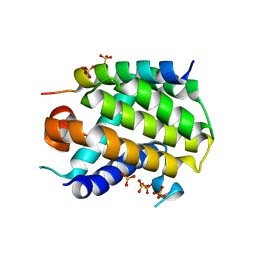 | |
6YLD
 
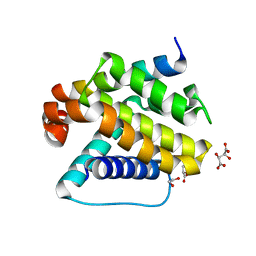 | | Crystal structure of Trichoplax adhaerens trBcl-2L2 bound to trBak BH3 | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2 homologous antagonist/killer, Bcl-2-like protein 1, ... | | Authors: | D Sa, J, Banjara, S, Kvansakul, M. | | Deposit date: | 2020-04-07 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Ancient and conserved functional interplay between Bcl-2 family proteins in the mitochondrial pathway of apoptosis.
Sci Adv, 6, 2020
|
|
6XY6
 
 | |
6XY4
 
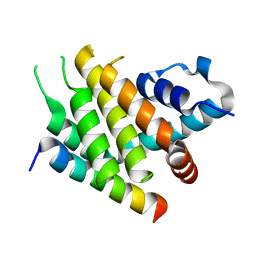 | |
5TZQ
 
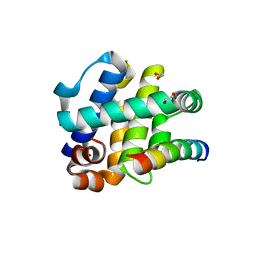 | | Crystal Structure of FPV039:Bmf BH3 complex | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2-like protein FPV039, Bcl-2-modifying factor | | Authors: | Anasir, M.I, Kvansakul, M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of apoptosis inhibition by the fowlpox virus protein FPV039.
J. Biol. Chem., 292, 2017
|
|
5TZP
 
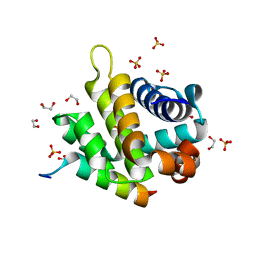 | | Crystal structure of FPV039:Bik BH3 complex | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2-like protein FPV039, Bik, ... | | Authors: | Anasir, M.I, Kvansakul, M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-05-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis of apoptosis inhibition by the fowlpox virus protein FPV039.
J. Biol. Chem., 292, 2017
|
|
5TWA
 
 | | Crystal structure of Geodia cydonium BHP2 in complex with Lubomirskia baicalensis Bak-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAK-2 protein, ... | | Authors: | Caria, S, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2016-11-12 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insight into an evolutionarily ancient programmed cell death regulator - the crystal structure of marine sponge BHP2 bound to LB-Bak-2.
Cell Death Dis, 8, 2017
|
|
5UA5
 
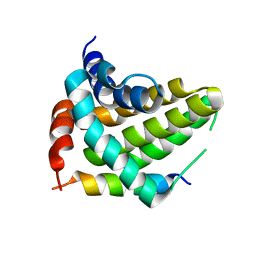 | |
5UA4
 
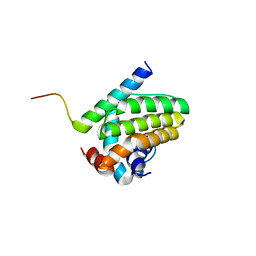 | |
5VWK
 
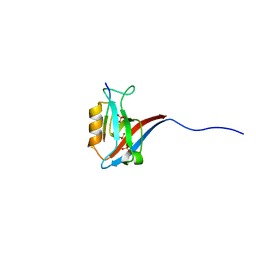 | | Crystal structure of human Scribble PDZ1:Beta-PIX complex | | Descriptor: | Beta-PIX, Protein scribble homolog, SULFATE ION | | Authors: | Lim, K.Y.B, Kvansakul, M. | | Deposit date: | 2017-05-22 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the differential interaction of Scribble PDZ domains with the guanine nucleotide exchange factor beta-PIX.
J. Biol. Chem., 292, 2017
|
|
5VMN
 
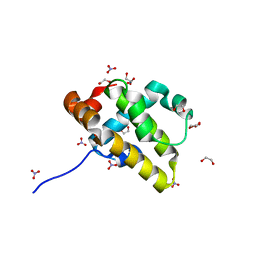 | | Crystal structure of grouper iridovirus GIV66 | | Descriptor: | 1,2-ETHANEDIOL, Bak protein, NITRATE ION | | Authors: | Banjara, S, Kvansakul, M. | | Deposit date: | 2017-04-27 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Grouper iridovirus GIV66 is a Bcl-2 protein that inhibits apoptosis by exclusively sequestering Bim.
J. Biol. Chem., 293, 2018
|
|
5VWI
 
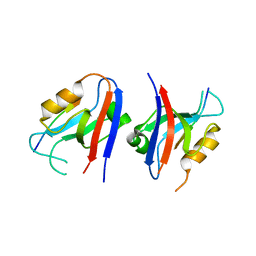 | |
5VWC
 
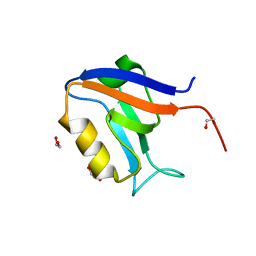 | | Crystal structure of human Scribble PDZ1 domain | | Descriptor: | 1,2-ETHANEDIOL, Protein scribble homolog | | Authors: | Lim, K.Y.B, Kvansakul, M. | | Deposit date: | 2017-05-21 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.911 Å) | | Cite: | Structural basis for the differential interaction of Scribble PDZ domains with the guanine nucleotide exchange factor beta-PIX.
J. Biol. Chem., 292, 2017
|
|
5VMO
 
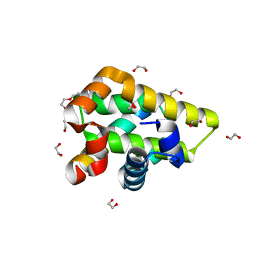 | | Crystal structure of grouper iridovirus GIV66:Bim complex | | Descriptor: | 1,2-ETHANEDIOL, Bak protein, Bcl-2 interacting mediator of cell death, ... | | Authors: | Banjara, S, Kvansakul, M. | | Deposit date: | 2017-04-28 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Grouper iridovirus GIV66 is a Bcl-2 protein that inhibits apoptosis by exclusively sequestering Bim.
J. Biol. Chem., 293, 2018
|
|
5WOS
 
 | |
