2HNI
 
 | |
1WP4
 
 | |
1X1E
 
 | |
1XKV
 
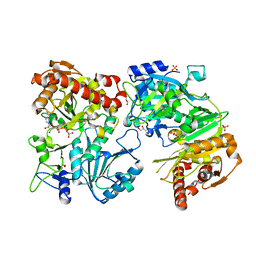 | | Crystal Structure Of ATP-Dependent Phosphoenolpyruvate Carboxykinase From Thermus thermophilus HB8 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent phosphoenolpyruvate carboxykinase, CALCIUM ION, ... | | Authors: | Sugahara, M, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-09-29 | | Release date: | 2005-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of ATP-dependent phosphoenolpyruvate carboxykinase from Thermus thermophilus HB8 showing the structural basis of induced fit and thermostability.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1X1O
 
 | |
1WXJ
 
 | |
1WZ8
 
 | |
1WN2
 
 | |
1WWK
 
 | |
1WM6
 
 | |
1WQ7
 
 | |
1X01
 
 | |
1WU8
 
 | |
1WS9
 
 | |
1WY0
 
 | |
1WY1
 
 | |
1WWZ
 
 | |
1WZN
 
 | |
1WXD
 
 | |
1X1Q
 
 | |
1WPY
 
 | |
1ZJJ
 
 | |
1YYA
 
 | |
1ZZG
 
 | |
2Z6R
 
 | | Crystal structure of Lys49 to Arg mutant of Diphthine synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Mizutani, H, Matsuura, Y, Krishna Swamy, B.S, Simanshu, D.K, Murthy, M.R.N, Kunishima, N. | | Deposit date: | 2007-08-08 | | Release date: | 2007-08-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of diphthine synthase from Pyrococcus horikoshii OT3
To be Published
|
|
