2EVB
 
 | |
1UB3
 
 | | Crystal Structure of Tetrameric Structure of Aldolase from thermus thermophilus HB8 | | Descriptor: | 1-HYDROXY-PENTANE-3,4-DIOL-5-PHOSPHATE, Aldolase protein | | Authors: | Lokanath, N.K, Miyano, M, Yokoyama, S, Kuramitsu, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-03-28 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of aldolase from Thermus thermophilus HB8 showing the contribution of oligomeric state to thermostability.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2CUN
 
 | |
2D8D
 
 | |
2D29
 
 | |
2D3K
 
 | |
2D4E
 
 | |
2D1Y
 
 | |
2D5C
 
 | | Crystal Structure of Shikimate 5-Dehydrogenase (AroE) from Thermus Thermophilus HB8 in complex with shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, SULFATE ION, shikimate 5-dehydrogenase | | Authors: | Bagautdinov, B, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-31 | | Release date: | 2006-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structures of Shikimate Dehydrogenase AroE from Thermus thermophilus HB8 and its Cofactor and Substrate Complexes: Insights into the Enzymatic Mechanism
J.Mol.Biol., 373, 2007
|
|
2D8A
 
 | |
2DCL
 
 | |
2DC4
 
 | |
2DEQ
 
 | |
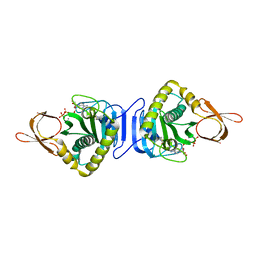
2DTI
 
 | |
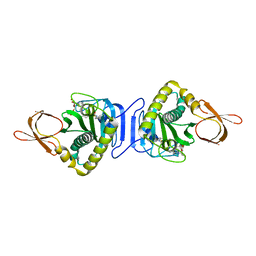
2DVE
 
 | |
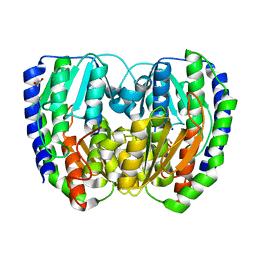
2DEC
 
 | |
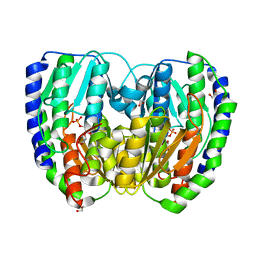
2DF8
 
 | |
2DR3
 
 | |
2DTH
 
 | |
2DPN
 
 | |
2DKG
 
 | |
2DTT
 
 | |
2DV3
 
 | |
2DFU
 
 | | Crystal structure of the 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase from Thermus Thermophilus HB8 | | Descriptor: | probable 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase | | Authors: | Mizutani, H, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-03 | | Release date: | 2006-09-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase from Thermus Thermophilus HB8
To be Published
|
|
2DPL
 
 | |
