2EGR
 
 | |
2EBB
 
 | |
2EBJ
 
 | | Crystal structure of pyrrolidone carboxyl peptidase from Thermus thermophilus | | Descriptor: | Pyrrolidone carboxyl peptidase | | Authors: | Kumarevel, T.S, Karthe, P, Agari, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-08 | | Release date: | 2007-08-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of pyrrolidone carboxyl peptidase from Thermus thermophilus
To be Published
|
|
2EEZ
 
 | |
2EHH
 
 | |
2EIU
 
 | | Crystal Structure of a Putative protein (AQ1627) from Aquifex aeolicus | | Descriptor: | Hypothetical protein aq_1627 | | Authors: | Kumarevel, T.S, Karthe, P, Nakano, N, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-13 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Putative protein (AQ1627) from Aquifex aeolicus
To be Published
|
|
2EGJ
 
 | |
2EHP
 
 | | Crystal Structure of a Putative protein (AQ1627) from Aquifex aeolicus | | Descriptor: | aq_1627 protein | | Authors: | Kumarevel, T.S, Karthe, P, Nakano, N, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-07 | | Release date: | 2007-09-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of a Putative protein (AQ1627) from Aquifex aeolicus
To be Published
|
|
2E8C
 
 | |
2EF4
 
 | |
2E8F
 
 | |
2EFO
 
 | | Crystal structure of Tyr77 to Ala of ST1022 from Sulfolobus tokodaii 7 | | Descriptor: | 150aa long hypothetical transcriptional regulator, MAGNESIUM ION | | Authors: | Kumarevel, T.S, Karthe, P, Nakano, N, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-23 | | Release date: | 2008-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of glutamine receptor protein from Sulfolobus tokodaii strain 7 in complex with its effector L-glutamine: implications of effector binding in molecular association and DNA binding.
Nucleic Acids Res., 36, 2008
|
|
2EFQ
 
 | | Crystal Structure of Thr134 to Ala of ST1022-Glutamine Complex from Sulfolobus tokodaii 7 | | Descriptor: | 150aa long hypothetical transcriptional regulator, GLUTAMINE, MAGNESIUM ION | | Authors: | Kumarevel, T.S, Karthe, P, Nakano, N, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-23 | | Release date: | 2008-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of glutamine receptor protein from Sulfolobus tokodaii strain 7 in complex with its effector L-glutamine: implications of effector binding in molecular association and DNA binding.
Nucleic Acids Res., 36, 2008
|
|
2EGI
 
 | |
2EBD
 
 | |
2E7W
 
 | | Crystal structure of the Lrp/AsnC like transcriptional regulators from Sulfolobus tokodaii 7 | | Descriptor: | 150aa long hypothetical transcriptional regulator, GLUTAMINE, MAGNESIUM ION | | Authors: | Kumarevel, T.S, Karthe, P, Nakano, N, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-15 | | Release date: | 2008-01-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of glutamine receptor protein from Sulfolobus tokodaii strain 7 in complex with its effector L-glutamine: implications of effector binding in molecular association and DNA binding.
Nucleic Acids Res., 36, 2008
|
|
2EBG
 
 | |
2PN6
 
 | | Crystal Structure of S32A of ST1022-Gln complex from Sulfolobus tokodaii | | Descriptor: | 150aa long hypothetical transcriptional regulator, GLUTAMINE, MAGNESIUM ION | | Authors: | Kumarevel, T.S, Karthe, P, Nakano, N, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-23 | | Release date: | 2008-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structure of glutamine receptor protein from Sulfolobus tokodaii strain 7 in complex with its effector L-glutamine: implications of effector binding in molecular association and DNA binding
Nucleic Acids Res., 36, 2008
|
|
2PMH
 
 | | Crystal structure of Thr132Ala of ST1022 from Sulfolobus tokodaii | | Descriptor: | 150aa long hypothetical transcriptional regulator, GLUTAMINE, MAGNESIUM ION, ... | | Authors: | Kumarevel, T.S, Karthe, P, Nakano, N, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-22 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of glutamine receptor protein from Sulfolobus tokodaii strain 7 in complex with its effector L-glutamine: implications of effector binding in molecular association and DNA binding
Nucleic Acids Res., 36, 2008
|
|
1CHV
 
 | |
1FMM
 
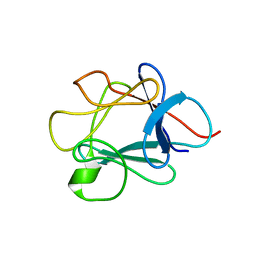 | | SOLUTION STRUCTURE OF NFGF-1 | | Descriptor: | ACIDIC FIBROBLAST GROWTH FACTOR | | Authors: | Arunkumar, A.I, Srisailam, S, Kumar, T.K.S, Chiu, I.M, Yu, C. | | Deposit date: | 2000-08-18 | | Release date: | 2001-08-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and stability of an acidic fibroblast growth factor from Notophthalmus viridescens.
J.Biol.Chem., 277, 2002
|
|
1HP9
 
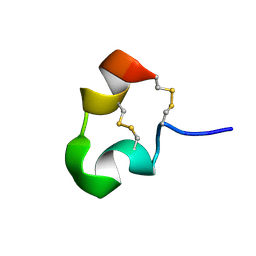 | | kappa-Hefutoxins: a novel Class of Potassium Channel Toxins from Scorpion venom | | Descriptor: | kappa-hefutoxin 1 | | Authors: | Srinivasan, K.N, Sivaraja, V, Huys, I, Sasaki, T, Cheng, B, Kumar, T.K.S, Sato, K, Tytgat, J, Yu, C, Brian Chia, C.S, Ranganathan, S, Bowie, J.H, Kini, R.M, Gopalakrishnakone, P. | | Deposit date: | 2000-12-12 | | Release date: | 2002-08-28 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | kappa-Hefutoxin1, a novel toxin from the scorpion Heterometrus fulvipes with unique structure and function. Importance of the functional diad in potassium channel selectivity.
J.Biol.Chem., 277, 2002
|
|
1KBT
 
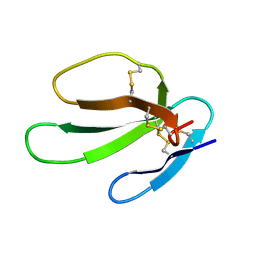 | | SOLUTION STRUCTURE OF CARDIOTOXIN IV, NMR, 12 STRUCTURES | | Descriptor: | CTX IV | | Authors: | Jeng, J.Y, Kumar, T.K.S, Jayaraman, G, Yu, C. | | Deposit date: | 1996-07-22 | | Release date: | 1997-07-23 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of the hemolytic activity and solution structures of two snake venom cardiotoxin analogues which only differ in their N-terminal amino acid.
Biochemistry, 36, 1997
|
|
1KBS
 
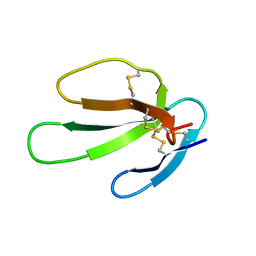 | | SOLUTION STRUCTURE OF CARDIOTOXIN IV, NMR, 1 STRUCTURE | | Descriptor: | CTX IV | | Authors: | Jeng, J.Y, Kumar, T.K.S, Jayaraman, G, Yu, C. | | Deposit date: | 1996-07-22 | | Release date: | 1997-07-23 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of the hemolytic activity and solution structures of two snake venom cardiotoxin analogues which only differ in their N-terminal amino acid.
Biochemistry, 36, 1997
|
|
1I9G
 
 | | CRYSTAL STRUCTURE OF AN ADOMET DEPENDENT METHYLTRANSFERASE | | Descriptor: | HYPOTHETICAL PROTEIN RV2118C, S-ADENOSYLMETHIONINE | | Authors: | Gupta, A, Kumar, P.H, Dineshkumar, T.K, Varshney, U, Subramanya, H.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2001-03-20 | | Release date: | 2001-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of Rv2118c: an AdoMet-dependent methyltransferase from Mycobacterium tuberculosis H37Rv.
J.Mol.Biol., 312, 2001
|
|
