1TXA
 
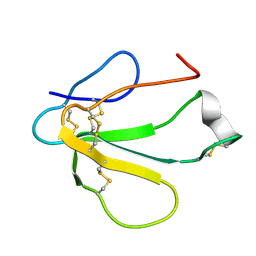 | | SOLUTION NMR STRUCTURE OF TOXIN B, A LONG NEUROTOXIN FROM THE VENOM OF THE KING COBRA, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TOXIN B | | Authors: | Peng, S.-S, Kumar, T.K.S, Jayaraman, G, Chang, C.-C, Yu, C. | | Deposit date: | 1996-07-20 | | Release date: | 1997-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of toxin b, a long neurotoxin from the venom of the king cobra (Ophiophagus hannah).
J.Biol.Chem., 272, 1997
|
|
1TXB
 
 | | SOLUTION NMR STRUCTURE OF TOXIN B, A LONG NEUROTOXIN FROM THE VENOM OF THE KING COBRA, 10 STRUCTURES | | Descriptor: | TOXIN B | | Authors: | Peng, S.-S, Kumar, T.K.S, Jayaraman, G, Chang, C.-C, Yu, C. | | Deposit date: | 1996-07-20 | | Release date: | 1997-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of toxin b, a long neurotoxin from the venom of the king cobra (Ophiophagus hannah).
J.Biol.Chem., 272, 1997
|
|
2CXJ
 
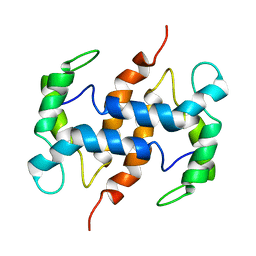 | |
3UV1
 
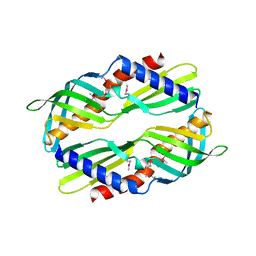 | | Crystal structure a major allergen from dust mite | | Descriptor: | Der f 7 allergen | | Authors: | Tan, K.W, Kumar, T, Chew, F.T, Mok, Y.K. | | Deposit date: | 2011-11-29 | | Release date: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Der f 7, a Dust Mite Allergen from Dermatophagoides farinae.
Plos One, 7, 2012
|
|
1X3P
 
 | |
1X32
 
 | |
1WVZ
 
 | |
1X3Q
 
 | |
2YSW
 
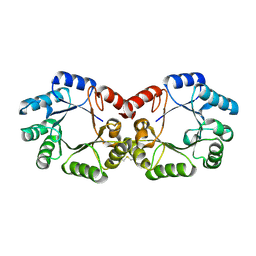 | | Crystal Structure of the 3-dehydroquinate dehydratase from Aquifex aeolicus VF5 | | Descriptor: | 3-dehydroquinate dehydratase | | Authors: | Tanaka, T, Kumarevel, T.S, Ebihara, A, Chen, L, Fu, Z.Q, Chrzas, J, Wang, B.C, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-04 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the 3-dehydroquinate dehydratase from Aquifex aeolicus VF5
To be Published
|
|
3X1U
 
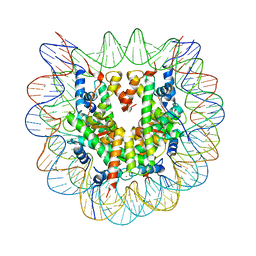 | |
3X1V
 
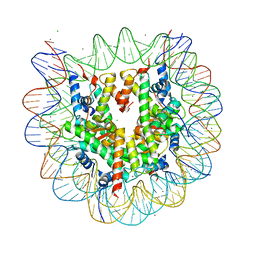 | |
3X1T
 
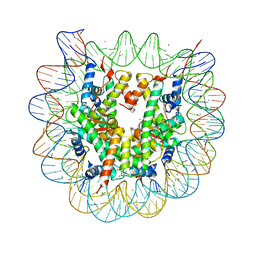 | |
3X1S
 
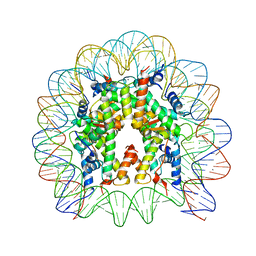 | | Crystal structure of the nucleosome core particle | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Sivaraman, P, Kumarevel, T.S. | | Deposit date: | 2014-11-27 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Structural and functional analyses of nucleosome complexes with mouse histone variants TH2a and TH2b, involved in reprogramming
Biochem.Biophys.Res.Commun., 464, 2015
|
|
3AOI
 
 | | RNA polymerase-Gfh1 complex (Crystal type 2) | | Descriptor: | Anti-cleavage anti-GreA transcription factor Gfh1, DNA (5'-D(*GP*GP*TP*CP*TP*GP*TP*AP*TP*CP*AP*CP*GP*AP*GP*CP*CP*A*CP*CP*GP*CP*CP*GP*CP*AP*T)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Tagami, S, Sekine, S, Kumarevel, T, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-09-30 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein
Nature, 468, 2010
|
|
4REU
 
 | | Revelation of Endogenously bound Fe2+ ions in the Crystal Structure of Ferritin from Escherichia coli | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FE (III) ION, ... | | Authors: | Thiruselvam, V, Ponnuswamy, M.N, Kumarevel, T.S. | | Deposit date: | 2014-09-24 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Revelation of endogenously bound Fe(2+) ions in the crystal structure of ferritin from Escherichia coli.
Biochem.Biophys.Res.Commun., 453, 2014
|
|
2YY6
 
 | |
3AOH
 
 | | RNA polymerase-Gfh1 complex (Crystal type 1) | | Descriptor: | Anti-cleavage anti-GreA transcription factor Gfh1, DNA (5'-D(*GP*GP*TP*CP*TP*GP*TP*AP*TP*CP*AP*CP*GP*AP*GP*CP*CP*AP*CP*CP*GP*CP*CP*GP*CP*AP*T)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Tagami, S, Sekine, S, Kumarevel, T, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-09-28 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein
Nature, 468, 2010
|
|
4OK9
 
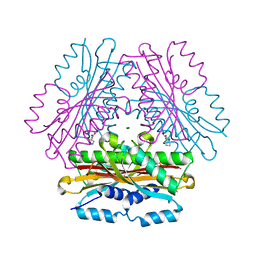 | |
3VNP
 
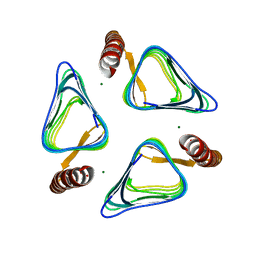 | | Crystal structure of hypothetical protein (GK2848) from Geobacillus Kaustophilus | | Descriptor: | ACETIC ACID, Hypothetical conserved protein, MAGNESIUM ION | | Authors: | Karthe, P.P, Kumarevel, T.S, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-01-17 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of hypothetical protein (GK2848) from Geobacillus Kaustophilus
To be Published
|
|
4OKQ
 
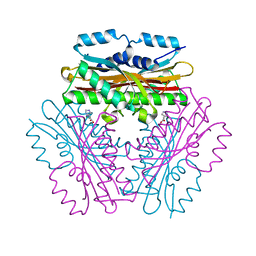 | |
2E85
 
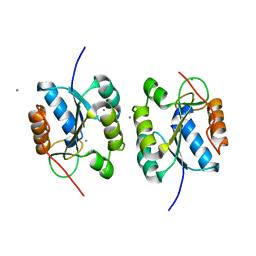 | | Crystal Structure of the Hydrogenase 3 Maturation protease | | Descriptor: | CALCIUM ION, Hydrogenase 3 maturation protease | | Authors: | Tanaka, T, Kumarevel, T.S, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-18 | | Release date: | 2008-01-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of hydrogenase maturating endopeptidase HycI from Escherichia coli
Biochem.Biophys.Res.Commun., 389, 2009
|
|
2DSI
 
 | | Crystal structure of Glu171 to Arg mutant of Diphthine synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Mizutani, H, Matsuura, Y, Saraboji, K, Malathy Sony, S.M, Ponnuswamy, M.N, Kumarevel, T.S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-06-30 | | Release date: | 2006-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of diphthine synthase from Pyrococcus horikoshii OT3
To be Published
|
|
3U6U
 
 | |
2EBY
 
 | | Crystal structure of a hypothetical protein from E. Coli | | Descriptor: | Putative HTH-type transcriptional regulator ybaQ, SULFATE ION | | Authors: | Karthe, P, Kumarevel, T.S, Ebihara, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-09 | | Release date: | 2007-08-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of a hypothetical protein from E. Coli
To be Published
|
|
2DSH
 
 | | Crystal structure of Lys26 to Tyr mutant of Diphthine synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Mizutani, H, Matsuura, Y, Saraboji, K, Malathy Sony, S.M, Ponnuswamy, M.N, Kumarevel, T.S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-06-30 | | Release date: | 2006-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of diphthine synthase from Pyrococcus horikoshii OT3
To be Published
|
|
