3MFY
 
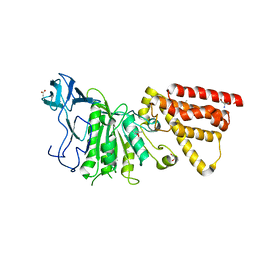 | | Structural characterization of the subunit A mutant F236A of the A-ATP synthase from Pyrococcus horikoshii | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | 著者 | Balakrishna, A.M, Kumar, A, Manimekali, M.S.S, Jeyakanthan, J, Gruber, G. | | 登録日 | 2010-04-05 | | 公開日 | 2010-07-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | The critical roles of residues P235 and F236 of subunit A of the motor protein A-ATP synthase in P-loop formation and nucleotide binding.
J.Mol.Biol., 401, 2010
|
|
4B2Z
 
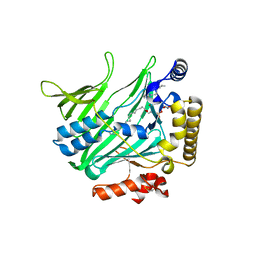 | | Structure of Osh6 in complex with phosphatidylserine | | 分子名称: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, ... | | 著者 | Maeda, K, Anand, K, Chiapparino, A, Kumar, A, Poletto, M, Kaksonen, M, Gavin, A.C. | | 登録日 | 2012-07-19 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Interactome Map Uncovers Phosphatidylserine Transport by Oxysterol-Binding Proteins
Nature, 501, 2013
|
|
4JC4
 
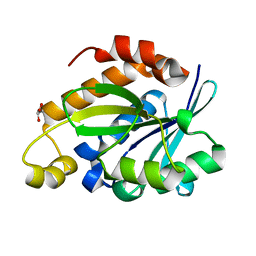 | | Crystal structure of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa at 2.25 angstrom resolution | | 分子名称: | GLYCEROL, Peptidyl-tRNA hydrolase | | 著者 | Singh, A, Kumar, A, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Arora, A, Singh, T.P. | | 登録日 | 2013-02-21 | | 公開日 | 2013-04-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural and binding studies of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa provide a platform for the structure-based inhibitor design against peptidyl-tRNA hydrolase
Biochem.J., 463, 2014
|
|
5GIV
 
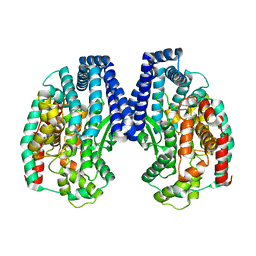 | | Crystal structure of M32 carboxypeptidase from Deinococcus radiodurans R1 | | 分子名称: | ACETATE ION, Carboxypeptidase 1, ZINC ION | | 著者 | Sharma, B, Singh, R, Yadav, P, Ghosh, B, Kumar, A, Jamdar, S.N, Makde, R.D. | | 登録日 | 2016-06-25 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Active site gate of M32 carboxypeptidases illuminated by crystal structure and molecular dynamics simulations
Biochim. Biophys. Acta, 1865, 2017
|
|
5GIU
 
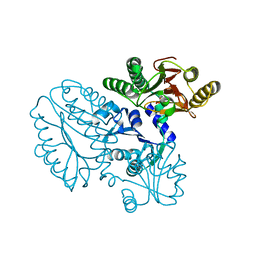 | | Crystal structure of Xaa-Pro peptidase from Deinococcus radiodurans, metal-free active site | | 分子名称: | PHOSPHATE ION, Proline dipeptidase, SODIUM ION | | 著者 | Are, V.N, Kumar, A, Singh, R, Ghosh, B, Jamdar, S.N, Makde, R.D. | | 登録日 | 2016-06-25 | | 公開日 | 2017-06-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Xaa-Pro peptidase from Deinococcus radiodurans, metal-free active site
To Be Published
|
|
5GJL
 
 | | Solution structure of SUMO from Plasmodium falciparum | | 分子名称: | Uncharacterized protein | | 著者 | Singh, J.S, Shukla, V.K, Gujrati, M, Mishra, R.K, Kumar, A. | | 登録日 | 2016-06-30 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure, dynamics and interaction study of SUMO from Plasmodium falciparum
To Be Published
|
|
8QOT
 
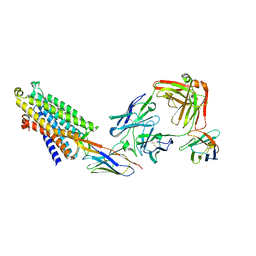 | | Structure of the mu opioid receptor bound to the antagonist nanobody NbE | | 分子名称: | Anti-Fab Nanobody, Mu-type opioid receptor, NabFab HC, ... | | 著者 | Yu, J, Kumar, A, Zhang, X, Martin, C, Raia, P, Manglik, A, Ballet, S, Boland, A, Stoeber, M. | | 登録日 | 2023-09-29 | | 公開日 | 2023-12-27 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural Basis of mu-Opioid Receptor-Targeting by a Nanobody Antagonist.
Biorxiv, 2023
|
|
5GIQ
 
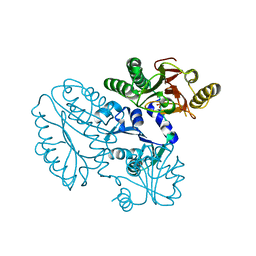 | | Xaa-Pro peptidase from Deinococcus radiodurans, Zinc bound | | 分子名称: | PHOSPHATE ION, Proline dipeptidase, ZINC ION | | 著者 | Are, V.N, Singh, R, Kumar, A, Ghosh, B, Jamdar, S.N, Makde, R.D. | | 登録日 | 2016-06-24 | | 公開日 | 2017-06-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structures and activities of widely conserved small prokaryotic aminopeptidases-P clarify classification of M24B peptidases.
Proteins, 2018
|
|
5I4F
 
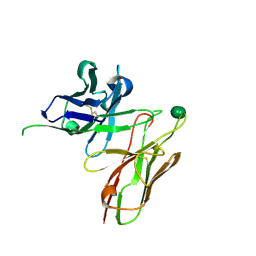 | | scFv 2D10 complexed with alpha 1,6 mannobiose | | 分子名称: | alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose, scFv 2D10 | | 著者 | Vashisht, S, Kumar, A, Kaur, K.J, Salunke, D.M. | | 登録日 | 2016-02-12 | | 公開日 | 2016-12-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.549 Å) | | 主引用文献 | Antibodies Can Exploit Molecular Crowding to Bind New Antigens at Noncanonical Paratope Positions
CHEMISTRYSELECT, 1, 2016
|
|
4QAJ
 
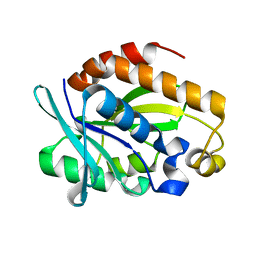 | | Crystal structure of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa at 1.5 Angstrom resolution | | 分子名称: | Peptidyl-tRNA hydrolase | | 著者 | Singh, A, Kumar, A, Gautam, L, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Arora, A, Singh, T.P. | | 登録日 | 2014-05-05 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural and binding studies of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa provide a platform for the structure-based inhibitor design against peptidyl-tRNA hydrolase
Biochem.J., 463, 2014
|
|
5X49
 
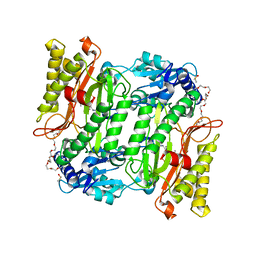 | | Crystal Structure of Human mitochondrial X-prolyl Aminopeptidase (XPNPEP3) | | 分子名称: | (2S,3R)-3-amino-2-hydroxy-4-phenylbutanoic acid, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | 著者 | Singh, R, Kumar, A, Ghosh, B, Jamdar, S, Makde, R.D. | | 登録日 | 2017-02-10 | | 公開日 | 2017-05-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structure of the human aminopeptidase XPNPEP3 and comparison of its in vitro activity with Icp55 orthologs: Insights into diverse cellular processes.
J. Biol. Chem., 292, 2017
|
|
5XEV
 
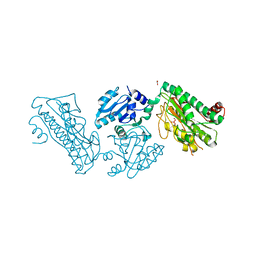 | | Crystal Structure of a novel Xaa-Pro dipeptidase from Deinococcus radiodurans | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | 著者 | Are, V.N, Kumar, A, Ghosh, B, Makde, R.D. | | 登録日 | 2017-04-06 | | 公開日 | 2017-10-18 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of a novel prolidase from Deinococcus radiodurans identifies new subfamily of bacterial prolidases.
Proteins, 85, 2017
|
|
5YZO
 
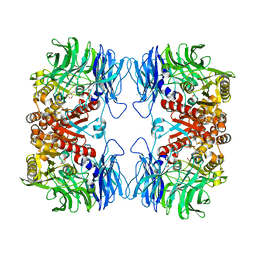 | | Crystal structure of S9 peptidase mutant (S514A) from Deinococcus radiodurans R1 | | 分子名称: | Acyl-peptide hydrolase, putative, DIMETHYL SULFOXIDE, ... | | 著者 | Yadav, P, Jamdar, S.N, Kumar, A, Ghosh, B, Makde, R.D. | | 登録日 | 2017-12-15 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
5YZN
 
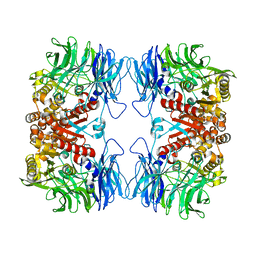 | | Crystal structure of S9 peptidase (active form) from Deinococcus radiodurans R1 | | 分子名称: | Acyl-peptide hydrolase, putative | | 著者 | Yadav, P, Jamdar, S.N, Kumar, A, Ghosh, B, Makde, R.D. | | 登録日 | 2017-12-15 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
5Z40
 
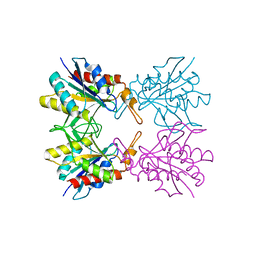 | |
5YZM
 
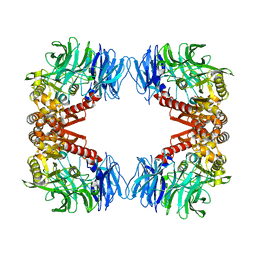 | | Crystal structure of S9 peptidase (inactive form) from Deinococcus radiodurans R1 | | 分子名称: | ACETATE ION, Acyl-peptide hydrolase, putative | | 著者 | Yadav, P, Jamdar, S.N, Kumar, A, Ghosh, B, Makde, R.D. | | 登録日 | 2017-12-15 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
6A4R
 
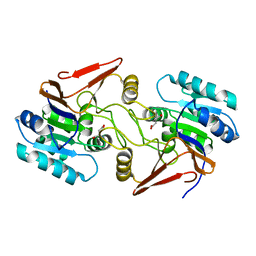 | | Crystal structure of aspartate bound peptidase E from Salmonella enterica | | 分子名称: | ASPARTIC ACID, Peptidase E | | 著者 | Yadav, P, Chandravanshi, K, Goyal, V.D, Singh, R, Kumar, A, Gokhale, S.M, Makde, R.D. | | 登録日 | 2018-06-20 | | 公開日 | 2018-10-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.828 Å) | | 主引用文献 | Structure of Asp-bound peptidase E from Salmonella enterica: Active site at dimer interface illuminates Asp recognition.
FEBS Lett., 592, 2018
|
|
6A4S
 
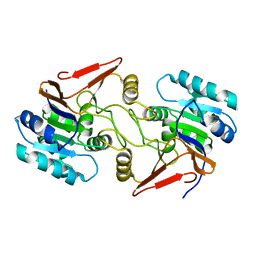 | | Crystal structure of peptidase E with ordered active site loop from Salmonella enterica | | 分子名称: | Peptidase E | | 著者 | Yadav, P, Chandravanshi, K, Goyal, V.D, Singh, R, Kumar, A, Gokhale, S.M, Makde, R.D. | | 登録日 | 2018-06-20 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of Asp-bound peptidase E from Salmonella enterica: Active site at dimer interface illuminates Asp recognition.
FEBS Lett., 592, 2018
|
|
6A9U
 
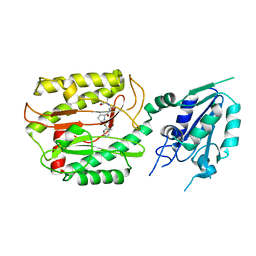 | | Crystal strcture of Icp55 from Saccharomyces cerevisiae bound to apstatin inhibitor | | 分子名称: | Intermediate cleaving peptidase 55, MANGANESE (II) ION, apstatin | | 著者 | Singh, R, Kumar, A, Goyal, V.D, Makde, R.D. | | 登録日 | 2018-07-16 | | 公開日 | 2019-01-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structures and biochemical analyses of intermediate cleavage peptidase: role of dynamics in enzymatic function.
FEBS Lett., 593, 2019
|
|
6A9V
 
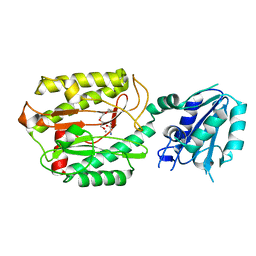 | | Crystal structure of Icp55 from Saccharomyces cerevisiae (N-terminal 42 residues deletion) | | 分子名称: | GLYCINE, Intermediate cleaving peptidase 55, MANGANESE (II) ION, ... | | 著者 | Singh, R, Kumar, A, Goyal, V.D, Makde, R.D. | | 登録日 | 2018-07-16 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structures and biochemical analyses of intermediate cleavage peptidase: role of dynamics in enzymatic function.
FEBS Lett., 593, 2019
|
|
6A4T
 
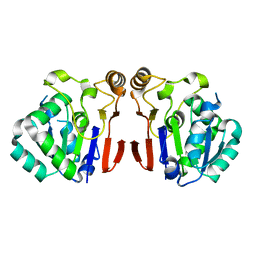 | | Crystal structure of Peptidase E from Deinococcus radiodurans R1 | | 分子名称: | Peptidase E | | 著者 | Yadav, P, Goyal, V.G, Kumar, A, Gokhale, S.M, Makde, R.D. | | 登録日 | 2018-06-20 | | 公開日 | 2019-06-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Catalytic triad heterogeneity in S51 peptidase family: Structural basis for functional variability.
Proteins, 87, 2019
|
|
6A9T
 
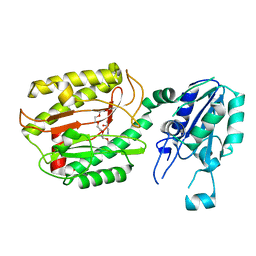 | | Crystal structure of Icp55 from Saccharomyces cerevisiae (N-terminal 58 residues deletion) | | 分子名称: | GLYCINE, Intermediate cleaving peptidase 55, MANGANESE (II) ION, ... | | 著者 | Singh, R, Kumar, A, Goyal, V.D, Makde, R.D. | | 登録日 | 2018-07-16 | | 公開日 | 2019-01-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structures and biochemical analyses of intermediate cleavage peptidase: role of dynamics in enzymatic function.
FEBS Lett., 593, 2019
|
|
7QV7
 
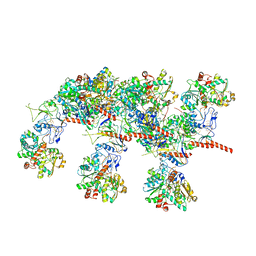 | | Cryo-EM structure of Hydrogen-dependent CO2 reductase. | | 分子名称: | Hydrogen dependent carbon dioxide reductase subunit FdhF, Hydrogen dependent carbon dioxide reductase subunit HycB3, Hydrogen dependent carbon dioxide reductase subunit HycB4, ... | | 著者 | Dietrich, H.M, Righetto, R.D, Kumar, A, Wietrzynski, W, Schuller, S.K, Trischler, R, Wagner, J, Schwarz, F.M, Engel, B.D, Mueller, V, Schuller, J.M. | | 登録日 | 2022-01-19 | | 公開日 | 2022-07-06 | | 最終更新日 | 2022-08-10 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Membrane-anchored HDCR nanowires drive hydrogen-powered CO 2 fixation.
Nature, 607, 2022
|
|
7RBF
 
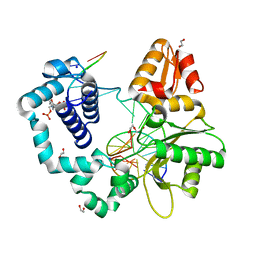 | | Human DNA polymerase beta crosslinked binary complex - B | | 分子名称: | 1,2-ETHANEDIOL, 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Reed, A.J, Kumar, A. | | 登録日 | 2021-07-06 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBG
 
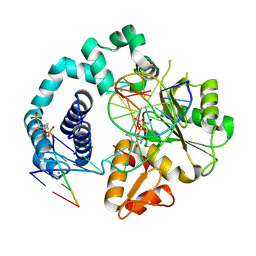 | | Human DNA polymerase beta crosslinked ternary complex 1 | | 分子名称: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, CALCIUM ION, ... | | 著者 | Reed, A.J, Kumar, A. | | 登録日 | 2021-07-06 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
