5CGA
 
 | | Structure of Hydroxyethylthiazole kinase ThiM from Staphylococcus aureus in complex with substrate analog 2-(1,3,5-trimethyl-1H-pyrazole-4-yl)ethanol | | Descriptor: | 2-(1,3,5-trimethyl-1H-pyrazol-4-yl)ethanol, Hydroxyethylthiazole kinase, MAGNESIUM ION | | Authors: | Kuenz, M, Drebes, J, Windshuegel, B, Cang, H, Wrenger, C, Betzel, C. | | Deposit date: | 2015-07-09 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of ThiM from Vitamin B1 biosynthetic pathway of Staphylococcus aureus - Insights into a novel pro-drug approach addressing MRSA infections.
Sci Rep, 6, 2016
|
|
5CGE
 
 | | Structure of Hydroxyethylthiazole Kinase ThiM from Staphylococcus aureus in complex with substrate analog 2-(2-methyl-1H-imidazole-1-yl)ethanol | | Descriptor: | 2-(2-methyl-1H-imidazol-1-yl)ethanol, Hydroxyethylthiazole kinase, MAGNESIUM ION | | Authors: | Kuenz, M, Drebes, J, Windshuegel, B, Cang, H, Wrenger, C, Betzel, C. | | Deposit date: | 2015-07-09 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure of ThiM from Vitamin B1 biosynthetic pathway of Staphylococcus aureus - Insights into a novel pro-drug approach addressing MRSA infections.
Sci Rep, 6, 2016
|
|
6H6T
 
 | |
5JKL
 
 | |
5JKM
 
 | |
5JKK
 
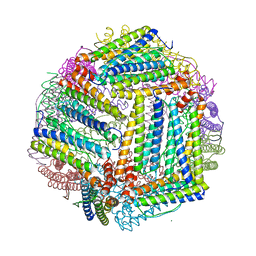 | |
6H6U
 
 | |
5COJ
 
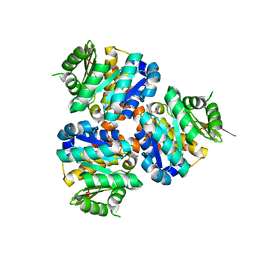 | | Structure of Hydroxyethylthiazole kinase ThiM from Staphylococcus aureus in complex with native substrate 2-(4-methyl-1,3-thiazol-5-yl)ethanol. | | Descriptor: | 2-(4-METHYL-THIAZOL-5-YL)-ETHANOL, Hydroxyethylthiazole kinase, MAGNESIUM ION | | Authors: | Drebes, J, Kuenz, M, Eberle, R.J, Oberthuer, D, Cang, H, Wrenger, C, Betzel, C. | | Deposit date: | 2015-07-20 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of ThiM from Vitamin B1 biosynthetic pathway of Staphylococcus aureus - Insights into a novel pro-drug approach addressing MRSA infections.
Sci Rep, 6, 2016
|
|
5CM5
 
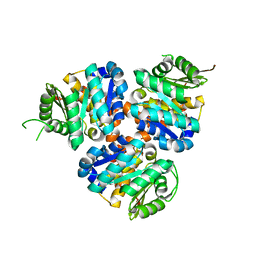 | | Structure of Hydroxyethylthiazole Kinase ThiM from Staphylococcus aureus | | Descriptor: | Hydroxyethylthiazole kinase | | Authors: | Drebes, J, Kuenz, M, Eberle, R.J, Oberthuer, D, Cang, H, Wrenger, C, Betzel, C. | | Deposit date: | 2015-07-16 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of ThiM from Vitamin B1 biosynthetic pathway of Staphylococcus aureus - Insights into a novel pro-drug approach addressing MRSA infections.
Sci Rep, 6, 2016
|
|
8QAQ
 
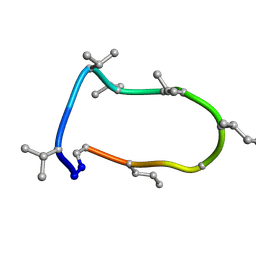 | | Conformations of macrocyclic peptides sampled by exact NOEs: models for cell-permeability. Conformation 1 of omphalotin A in apolar solvents. | | Descriptor: | TRP-MVA-ILE-MVA-MVA-SAR-MVA-IML-SAR-VAL-IML-SAR | | Authors: | Ruedisser, S.H, Matabaro, E, Sonderegger, L, Guentert, P, Kuenzler, M, Gossert, A.D. | | Deposit date: | 2023-08-23 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | SOLUTION NMR | | Cite: | Conformations of Macrocyclic Peptides Sampled by Nuclear Magnetic Resonance: Models for Cell-Permeability.
J.Am.Chem.Soc., 145, 2023
|
|
8Q7J
 
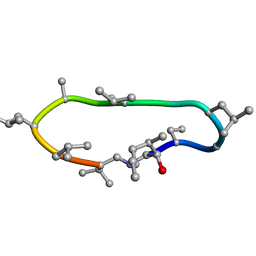 | | Conformations of macrocyclic peptides sampled by exact NOEs: models for cell-permeability | | Descriptor: | CYCLOSPORIN A | | Authors: | Ruedisser, S.H, Matabaro, E, Sonderegger, L, Guentert, P, Kuenzler, M, Gossert, A.D. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | SOLUTION NMR | | Cite: | Conformations of Macrocyclic Peptides Sampled by Nuclear Magnetic Resonance: Models for Cell-Permeability.
J.Am.Chem.Soc., 145, 2023
|
|
8QAS
 
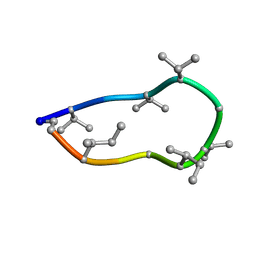 | | Conformations of macrocyclic peptides sampled by exact NOEs: models for cell-permeability. NMR structure of Omphalotin A in methanol / water indoleOut conformation. | | Descriptor: | TRP-MVA-ILE-MVA-MVA-SAR-MVA-IML-SAR-VAL-IML-SAR | | Authors: | Ruedisser, S.H, Matabaro, E, Sonderegger, L, Guentert, P, Kuenzler, M, Gossert, A.D. | | Deposit date: | 2023-08-23 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | SOLUTION NMR | | Cite: | Conformations of Macrocyclic Peptides Sampled by Nuclear Magnetic Resonance: Models for Cell-Permeability.
J.Am.Chem.Soc., 145, 2023
|
|
8QBP
 
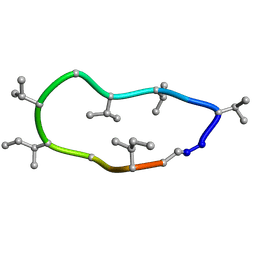 | | Conformations of macrocyclic peptides sampled by exact NOEs: models for cell-permeability. NMR structure of Omphalotin A in methanol / water indoleOut conformation. | | Descriptor: | TRP-MVA-ILE-MVA-MVA-SAR-MVA-IML-SAR-VAL-IML-SAR | | Authors: | Ruedisser, S.H, Matabaro, E, Sonderegger, L, Guentert, P, Kuenzler, M, Gossert, A.D. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Conformations of Macrocyclic Peptides Sampled by Nuclear Magnetic Resonance: Models for Cell-Permeability.
J.Am.Chem.Soc., 145, 2023
|
|
6ZRW
 
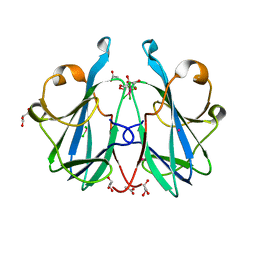 | | Crystal structure of the fungal lectin CML1 | | Descriptor: | ACETATE ION, GLYCEROL, Mucin-binding lectin 1, ... | | Authors: | Bleuler-Martinez, S, Olieric, V, Sharpe, M, Capitani, G, Aebi, M, Kuenzler, M. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-function relationship of a novel fucoside-binding fruiting body lectin from Coprinopsis cinerea exhibiting nematotoxic activity.
Glycobiology, 32, 2022
|
|
3N0K
 
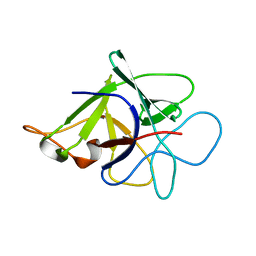 | | Proteinase inhibitor from Coprinopsis cinerea | | Descriptor: | Serine protease inhibitor 1 | | Authors: | Renko, M, Sabotic, J, Bleuler-Martinez, S, Kallert, S, Avanzo, P, Kos, J, Aebi, M, Kuenzler, M, Turk, D. | | Deposit date: | 2010-05-14 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Trypsin Inhibition and Entomotoxicity of Cospin, Serine Protease Inhibitor Involved in Defense of Coprinopsis cinerea Fruiting Bodies.
J.Biol.Chem., 287, 2012
|
|
2WKK
 
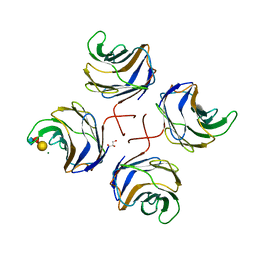 | | Identification of the glycan target of the nematotoxic fungal galectin CGL2 in Caenorhabditis elegans | | Descriptor: | GALECTIN-2, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Butschi, A, Titz, A, Waelti, M, Olieric, V, Paschinger, K, Xiaoqiang, G, Seeberger, P.H, Wilson, I.B.H, Aebi, M, Hengartner, M.O, Kuenzler, M. | | Deposit date: | 2009-06-14 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Caenorhabditis Elegans N-Glycan Core Beta-Galactoside Confers Sensitivity Towards Nematotoxic Fungal Galectin Cgl2.
Plos Pathog., 6, 2010
|
|
2LIQ
 
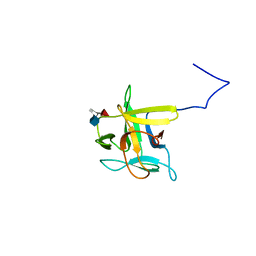 | | Solution structure of CCL2 in complex with glycan | | Descriptor: | CCL2 lectin, alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside | | Authors: | Schubert, M, Bleuler-Martinez, S, Walti, M.A, Egloff, P, Aebi, M, Kuenzler, M, Allain, F.H.-T. | | Deposit date: | 2011-08-30 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Plasticity of the beta-Trefoil Protein Fold in the Recognition and Control of Invertebrate Predators and Parasites by a Fungal Defence System
Plos Pathog., 8, 2012
|
|
2R0H
 
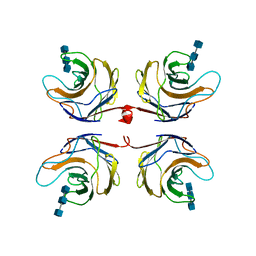 | | Fungal lectin CGL3 in complex with chitotriose (chitotetraose) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CGL3 lectin | | Authors: | Waelti, M.A, Walser, P.J, Thore, S, Gruenler, A, Ban, N, Kuenzler, M, Aebi, M. | | Deposit date: | 2007-08-20 | | Release date: | 2008-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Chitotetraose Coordination by CGL3, a Novel Galectin-Related Protein from Coprinopsis cinerea
J.Mol.Biol., 379, 2008
|
|
2R0F
 
 | | Ligand free structure of fungal lectin CGL3 | | Descriptor: | CGL3 lectin | | Authors: | Waelti, M.A, Walser, P.J, Thore, S, Gruenler, A, Ban, N, Kuenzler, M, Aebi, M. | | Deposit date: | 2007-08-19 | | Release date: | 2008-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Chitotetraose Coordination by CGL3, a Novel Galectin-Related Protein from Coprinopsis cinerea
J.Mol.Biol., 379, 2008
|
|
1ULC
 
 | | CGL2 in complex with lactose | | Descriptor: | beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
1ULF
 
 | | CGL2 in complex with Blood Group A tetrasaccharide | | Descriptor: | alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
1ULE
 
 | | CGL2 in complex with linear B2 trisaccharide | | Descriptor: | alpha-D-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
1ULD
 
 | | CGL2 in complex with blood group H type II | | Descriptor: | alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
1ULG
 
 | | CGL2 in complex with Thomsen-Friedenreich antigen | | Descriptor: | beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
1UL9
 
 | | CGL2 ligandfree | | Descriptor: | galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
