5S70
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with EN300-181428 | | Descriptor: | (5R)-2-methyl-4,5,6,7-tetrahydro-1H-benzimidazol-5-amine, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2020-11-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.327 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5A7P
 
 | | Crystal structure of human JMJD2A in complex with compound 36 | | Descriptor: | 1,2-ETHANEDIOL, 2-[5-[(5-methyl-1,2-oxazol-3-yl)carbonylamino]-2-oxidanyl-phenyl]pyridine-4-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Nowak, R, Velupillai, S, Krojer, T, Gileadi, C, Johansson, C, Korczynska, M, Le, D.D, Younger, N, Gregori-Puigjane, E, Tumber, A, Iwasa, E, Pollock, S.B, Ortiz Torres, I, Kopec, J, Tallant, C, Froese, S, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Shoichet, B.K, Fujimori, D.G, Oppermann, U. | | Deposit date: | 2015-07-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Docking and Linking of Fragments to Discover Jumonji Histone Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5SAF
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with EN300-321461 | | Descriptor: | 6,7-dihydro-5H-pyrrolo[2,3-d]pyrimidine, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
7B6G
 
 | | Crystal structure of MurE from E.coli in complex with Z1675346324 | | Descriptor: | DIMETHYL SULFOXIDE, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2,6-diaminopimelate ligase, trans-3-[(2,6-dimethylpyrimidin-4-yl)(methyl)amino]cyclobutan-1-ol | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
7B6Q
 
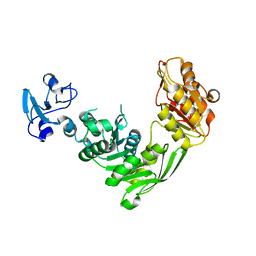 | | Crystal structure of MurE from E.coli in complex with Z57299526 | | Descriptor: | ISOPROPYL ALCOHOL, N-[(furan-2-yl)methyl]-1H-benzimidazol-2-amine, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-08 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
5S8X
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with starting material | | Descriptor: | 3,4-dichlorobenzoate, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassel-Hart , S, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structures of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2 soaked with crude reaction mixtures
To Be Published
|
|
5SAI
 
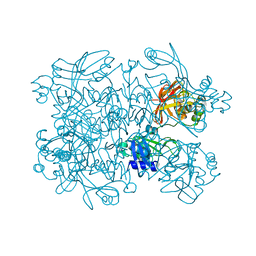 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z1424343998 | | Descriptor: | N-{2-[(propan-2-yl)sulfanyl]phenyl}urea, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.022 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
7B53
 
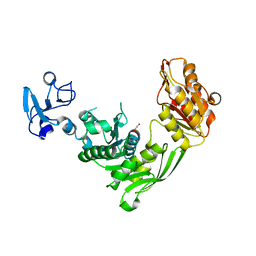 | | Crystal structure of MurE from E.coli | | Descriptor: | 1,2-ETHANEDIOL, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
5S96
 
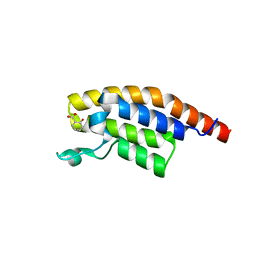 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with starting material | | Descriptor: | 2-HYDROXYBENZOIC ACID, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassel-Hart , S, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Crystal Structures of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2 soaked with crude reaction mixtures
To Be Published
|
|
5SAC
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z59181945 | | Descriptor: | CITRIC ACID, N-hydroxyquinoline-2-carboxamide, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5S98
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with starting material | | Descriptor: | 5-bromopyridine-3-carboxylic acid, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassel-Hart , S, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Structures of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2 soaked with crude reaction mixtures
To Be Published
|
|
5SA5
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z1530301542 | | Descriptor: | 4-ethyl-2-(1H-imidazol-5-yl)-1,3-thiazole, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SA7
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z1673618163 | | Descriptor: | 4-amino-N-(2-hydroxyethyl)-N-methylbenzene-1-sulfonamide, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.222 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5S99
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with starting material | | Descriptor: | 2,3-dihydrothieno[3,4-b][1,4]dioxine-5-carboxylic acid, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassel-Hart , S, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystal Structures of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2 soaked with crude reaction mixtures
To Be Published
|
|
5SAG
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with EN300-1605072 | | Descriptor: | 3-(1H-imidazol-2-yl)propan-1-amine, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5S9B
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex withstarting material | | Descriptor: | 3-methoxybenzoic acid, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassel-Hart , S, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.153 Å) | | Cite: | Crystal Structures of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2 soaked with crude reaction mixtures
To Be Published
|
|
5SAA
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z319891284 | | Descriptor: | 3-[(2S)-1-(methanesulfonyl)pyrrolidin-2-yl]-5-methyl-1,2-oxazole, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.239 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5S9G
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with starting material | | Descriptor: | 1,3-benzothiazole-6-carboxylic acid, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassel-Hart , S, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.091 Å) | | Cite: | Crystal Structures of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2 soaked with crude reaction mixtures
To Be Published
|
|
5S9J
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with starting material | | Descriptor: | 2-azanyl-3-methyl-benzoic acid, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassel-Hart , S, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structures of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2 soaked with crude reaction mixtures
To Be Published
|
|
5SA4
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z239136710 | | Descriptor: | N-(5-methyl-1H-pyrazol-3-yl)acetamide, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.046 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SA8
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z68299550 | | Descriptor: | 3-chloro-N-(1-hydroxy-2-methylpropan-2-yl)benzamide, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SAD
 
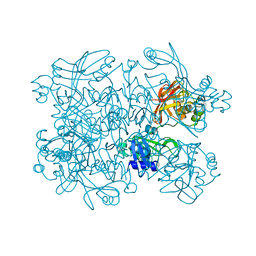 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z425449682 | | Descriptor: | (3-phenyl-1,2-oxazol-5-yl)methylazanium, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
7B6M
 
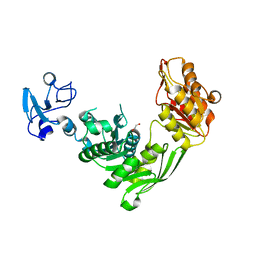 | | Crystal structure of MurE from E.coli in complex with Z1198948504 | | Descriptor: | 1,2-ETHANEDIOL, N-[2-(2,5-Dioxopyrrolidin-1-yl)ethyl]-3-methylbenzamide, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC), Structural Genomics Consortium for Research on Gene Expression (SGCGES) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
5SA9
 
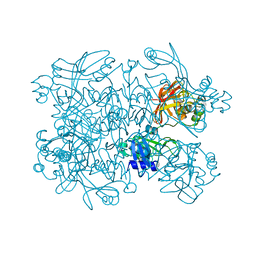 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z2697514548 | | Descriptor: | 1-methylindazole-3-carboxamide, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
7B60
 
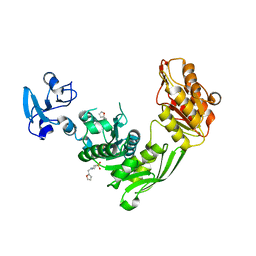 | | Crystal structure of MurE from E.coli in complex with Z1269139261 | | Descriptor: | 4-[(furan-2-yl)methyl]-1lambda~6~,4-thiazinane-1,1-dione, DIMETHYL SULFOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
