3FRP
 
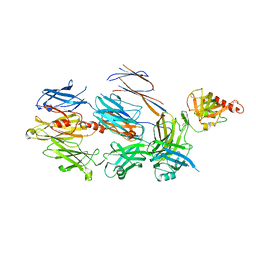 | | Crystal Structure of Cobra Venom Factor, a Co-factor for C3- and C5 convertase CVFBb | | Descriptor: | CALCIUM ION, Cobra venom factor alpha chain, Cobra venom factor beta chain, ... | | Authors: | Krishnan, V, Narayana, S.V.L. | | Deposit date: | 2009-01-08 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The Crystal Structure of Cobra Venom Factor, a Cofactor for C3- and C5-Convertase CVFBb.
Structure, 17, 2009
|
|
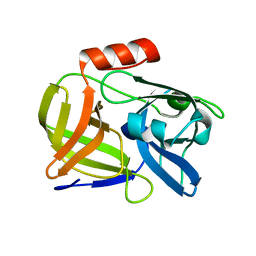
4JCN
 
 | |
3TW0
 
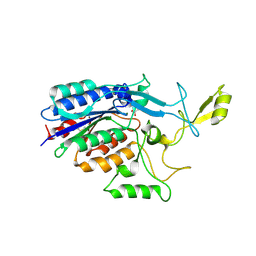 | |
3TXA
 
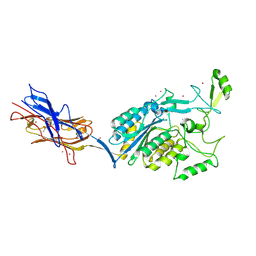 | | Structural Analysis of Adhesive Tip pilin, GBS104 from Group B Streptococcus agalactiae | | Descriptor: | CADMIUM ION, Cell wall surface anchor family protein, LITHIUM ION, ... | | Authors: | Krishnan, V, Narayana, S.V.L. | | Deposit date: | 2011-09-23 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.619 Å) | | Cite: | Structure of Streptococcus agalactiae tip pilin GBS104: a model for GBS pili assembly and host interactions.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3TVY
 
 | |
1HST
 
 | | CRYSTAL STRUCTURE OF GLOBULAR DOMAIN OF HISTONE H5 AND ITS IMPLICATIONS FOR NUCLEOSOME BINDING | | Descriptor: | HISTONE H5 | | Authors: | Ramakrishnan, V, Finch, J.T, Graziano, V, Lee, P.L, Sweet, R.M. | | Deposit date: | 1993-03-30 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of globular domain of histone H5 and its implications for nucleosome binding.
Nature, 362, 1993
|
|
1PKP
 
 | |
6M48
 
 | | Crystal structure of pilus adhesin, SpaC from Lactobacillus rhamnosus GG - P21212 form | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SpaC | | Authors: | Kant, A, Palva, A, Von Ossowaski, I, Krishnan, V. | | Deposit date: | 2020-03-05 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of lactobacillar SpaC reveals an atypical five-domain pilus tip adhesin: Exposing its substrate-binding and assembly in SpaCBA pili.
J.Struct.Biol., 211, 2020
|
|
7D41
 
 | |
5HTS
 
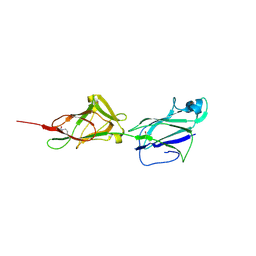 | | Crystal structure of shaft pilin spaA from Lactobacillus rhamnosus GG - D295N mutant | | Descriptor: | Cell surface protein SpaA | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2016-01-27 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
8WB8
 
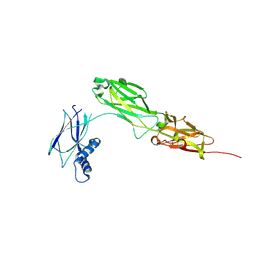 | |
8W5B
 
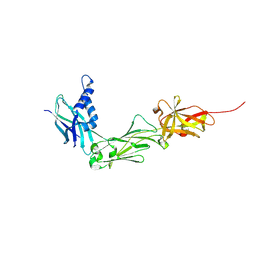 | | Crystal Structure of the shaft pilin LrpA from Ligilactobacillus ruminis | | Descriptor: | IODIDE ION, LPXTG-motif cell wall anchor domain protein | | Authors: | Prajapati, A, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2023-08-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The crystal structure of the N-terminal domain of the backbone pilin LrpA reveals a new closure-and-twist motion for assembling dynamic pili in Ligilactobacillus ruminis.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
5J4M
 
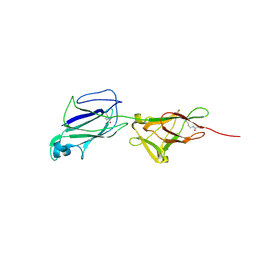 | | Crystal structure of shaft pilin SpaA from Lactobacillus rhamnosus GG - E269A/D295N double mutant | | Descriptor: | Cell surface protein SpaA | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2016-04-01 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
8GR6
 
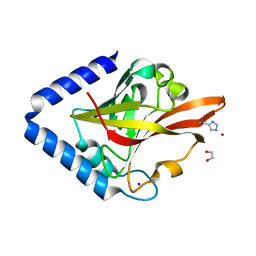 | | Crystal Structure of pilus-specific Sortase C from Streptococcus sanguinis | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Sortase-like protein, ... | | Authors: | Yadav, S, Parijat, P, Krishnan, V. | | Deposit date: | 2022-09-01 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of the pilus-specific sortase from early colonizing oral Streptococcus sanguinis captures an active open-lid conformation.
Int.J.Biol.Macromol., 243, 2023
|
|
8GMX
 
 | |
6M3Y
 
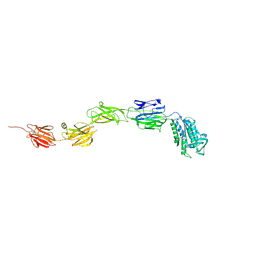 | | Crystal structure of pilus adhesin, SpaC from Lactobacillus rhamnosus GG - open conformation | | Descriptor: | MAGNESIUM ION, Pilus assembly protein | | Authors: | Kant, A, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2020-03-04 | | Release date: | 2020-07-29 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of lactobacillar SpaC reveals an atypical five-domain pilus tip adhesin: Exposing its substrate-binding and assembly in SpaCBA pili.
J.Struct.Biol., 211, 2020
|
|
6M7C
 
 | |
7W7I
 
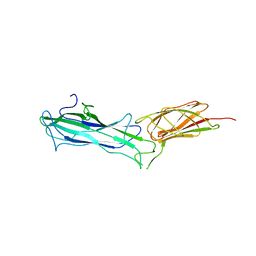 | |
7W6B
 
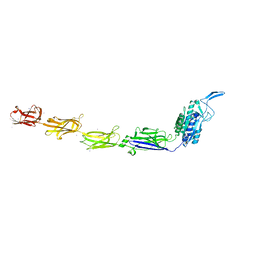 | |
3PHS
 
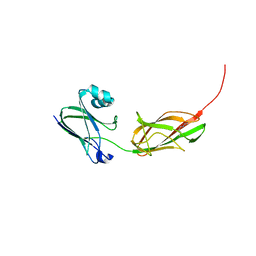 | |
3QDH
 
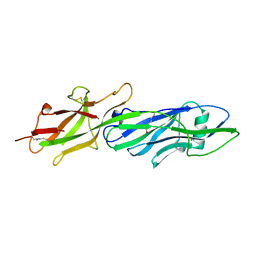 | |
8KB2
 
 | | Crystal Structure of M- and C-Domains of the shaft pilin LrpA from Ligilactobacillus ruminis - iodide derivative | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Prajapati, A, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2023-08-03 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The crystal structure of the N-terminal domain of the backbone pilin LrpA reveals a new closure-and-twist motion for assembling dynamic pili in Ligilactobacillus ruminis.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8KG4
 
 | | Crystal Structure of M- and C-Domains of the shaft pilin LrpA from Ligilactobacillus ruminis - orthorhombic form | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, IODIDE ION, ... | | Authors: | Prajapati, A, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2023-08-17 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The crystal structure of the N-terminal domain of the backbone pilin LrpA reveals a new closure-and-twist motion for assembling dynamic pili in Ligilactobacillus ruminis.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8KCL
 
 | |
5YU5
 
 | | Crystal structure of shaft pilin spaD from Lactobacillus rhamnosus GG | | Descriptor: | Pilus assembly protein | | Authors: | Chaurasia, P, Pratap, S, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2017-11-20 | | Release date: | 2018-06-20 | | Last modified: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Bent conformation of a backbone pilin N-terminal domain supports a three-stage pilus assembly mechanism.
Commun Biol, 1, 2018
|
|
