1OU0
 
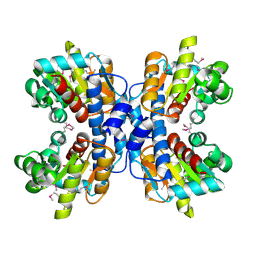 | | precorrin-8X methylmutase related protein | | Descriptor: | precorrin-8X methylmutase related protein | | Authors: | Cuff, M.E, Joachimiak, A, Korolev, S, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-03-24 | | Release date: | 2003-10-07 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a predicted precorrin-8x methylmutase from Thermoplasma acidophilum.
Proteins, 58, 2004
|
|
1KA8
 
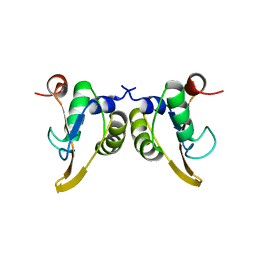 | | Crystal Structure of the Phage P4 Origin-Binding Domain | | Descriptor: | putative P4-specific DNA primase | | Authors: | Yeo, H.J, Ziegelin, G, Korolev, S, Calendar, R, Lanka, E, Waksman, G. | | Deposit date: | 2001-10-31 | | Release date: | 2002-04-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Phage P4 origin-binding domain structure reveals a mechanism for regulation of DNA-binding activity by homo- and heterodimerization of winged helix proteins.
Mol.Microbiol., 43, 2002
|
|
1L1O
 
 | | Structure of the human Replication Protein A (RPA) trimerization core | | Descriptor: | Replication protein A 14 kDa subunit, Replication protein A 32 kDa subunit, Replication protein A 70 kDa DNA-binding subunit, ... | | Authors: | Bochkareva, E.V, Korolev, S, Lees-Miller, S.P, Bochkarev, A. | | Deposit date: | 2002-02-19 | | Release date: | 2002-06-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA.
EMBO J., 21, 2002
|
|
1KJN
 
 | |
1P8C
 
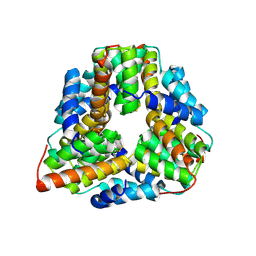 | | Crystal structure of TM1620 (APC4843) from Thermotoga maritima | | Descriptor: | conserved hypothetical protein | | Authors: | Kim, Y, Joachimiak, A, Brunzelle, J.S, Korolev, S.V, Edwards, A, Xu, X, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-05-06 | | Release date: | 2003-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure Analysis of Thermotoga maritima protein TM1620 (APC4843)
To be Published
|
|
1NC5
 
 | | Structure of Protein of Unknown Function of YteR from Bacillus Subtilis | | Descriptor: | hypothetical protein yTER | | Authors: | Zhang, R, Lozondra, L, Korolev, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-04 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.6 A crystal structure of YteR protein from Bacillus subtilis, a predicted lyase.
Proteins, 60, 2005
|
|
1NSL
 
 | | Crystal structure of Probable acetyltransferase | | Descriptor: | CHLORIDE ION, Probable acetyltransferase | | Authors: | Brunzelle, J.S, Korolev, S.V, Wu, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-27 | | Release date: | 2003-07-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Bacillus subtilis YdaF protein: A putative ribosomal N-acetyltransferase
Proteins, 57, 2004
|
|
