8R5J
 
 | | Crystal structure of MERS-CoV main protease | | Descriptor: | Non-structural protein 11 | | Authors: | Balcomb, B.H, Fairhead, M, Koekemoer, L, Lithgo, R.M, Aschenbrenner, J.C, Chandran, A.V, Godoy, A.S, Lukacik, P, Marples, P.G, Mazzorana, M, Ni, X, Strain-Damerell, C, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-11-16 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal structure of MERS-CoV main protease
To Be Published
|
|
8UM3
 
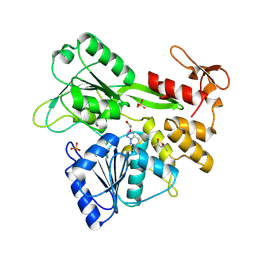 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z203039992 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 6-chlorotetrazolo[1,5-b]pyridazine, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-10-17 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z203039992
To Be Published
|
|
5SDR
 
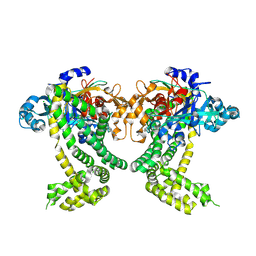 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z1273312153 | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION, N-methyl-1H-indole-7-carboxamide | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.077 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDF
 
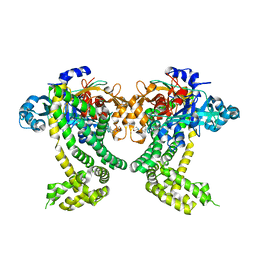 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z2856434834 | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION, N-(2,3-dimethylphenyl)-2-(morpholin-4-yl)acetamide | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (1.877 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDN
 
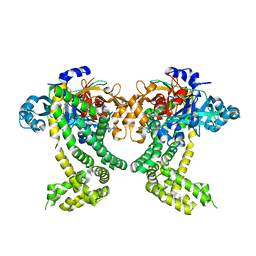 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z437584380 | | Descriptor: | (4-chlorophenyl)(thiomorpholin-4-yl)methanone, Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.023 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDD
 
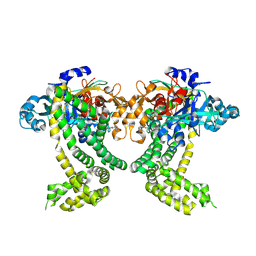 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z2856434879 | | Descriptor: | 2-[(4-methyl-1H-imidazol-5-yl)methyl]-1,2,3,4-tetrahydroisoquinoline, Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDM
 
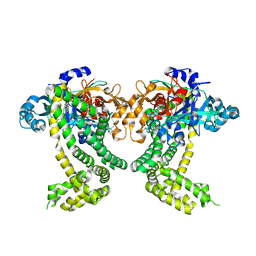 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z1328078283 | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION, N-(cyclopropylmethyl)-2,2,3,3-tetramethylazetidine-1-carboxamide | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDS
 
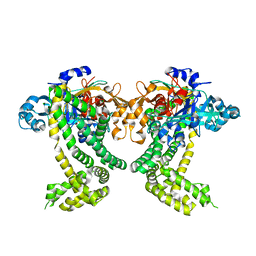 | | PanDDA analysis group deposition of ground-state model of Porphyromonas gingivalis DPP11 | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
8CO8
 
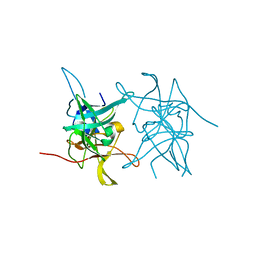 | | Structure of West Nile Virus NS2B-NS3 protease | | Descriptor: | Serine protease subunit NS2B,Serine protease/Helicase NS3 | | Authors: | Fairhead, M, Godoy, A.S, Koekemoer, L, Balcomb, B.H, Lithgo, R.M, Aschenbrenner, J.C, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-02-27 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of West Nile Virus NS2B-NS3 protease - to be published
To Be Published
|
|
8V7R
 
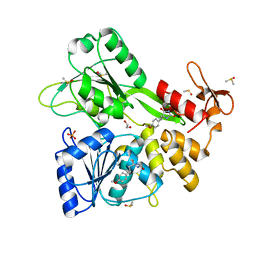 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z56772132 | | Descriptor: | (5R)-5-[2-(4-methoxyphenyl)ethyl]-5-methylimidazolidine-2,4-dione, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-12-04 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z56772132
To Be Published
|
|
8V7U
 
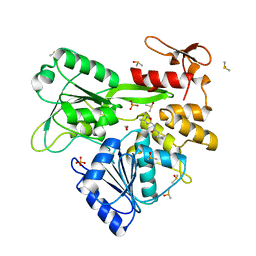 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z729726784 | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclopentyl-N-(3-methyl-1,2,4-oxadiazol-5-yl)acetamide, DIMETHYL SULFOXIDE, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-12-04 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z729726784
To Be Published
|
|
8BW4
 
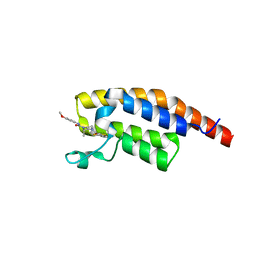 | | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative | | Descriptor: | (2R)-4-(3-fluoranylthiophen-2-yl)carbonyl-N-(4-methoxyphenyl)-2-methyl-piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassell-Hart, S, Bradshaw, W.J, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2022-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative
To Be Published
|
|
8BW3
 
 | | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative | | Descriptor: | (2S)-N-(cyclopropylmethyl)-2-methyl-4-(1-methyl-1H-pyrrole-2-carbonyl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassell-Hart, S, Bradshaw, W.J, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2022-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative
To Be Published
|
|
8BW2
 
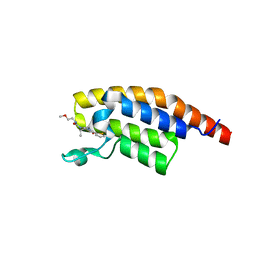 | | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative | | Descriptor: | (2R)-N-(2-methoxyethyl)-2-methyl-4-thiophen-2-ylcarbonyl-piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassell-Hart, S, Bradshaw, W.J, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2022-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative
To Be Published
|
|
6HZJ
 
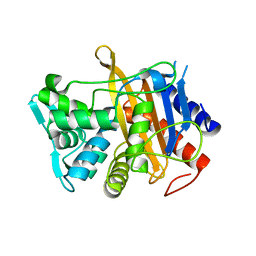 | | Apo structure of TP domain from clinical penicillin-resistant mutant Neisseria gonorrhoea strain 6140 Penicillin-Binding Protein 2 (PBP2) | | Descriptor: | Probable peptidoglycan D,D-transpeptidase PenA | | Authors: | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | Deposit date: | 2018-10-23 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Novel and Improved Crystal Structures of H. influenzae, E. coli and P. aeruginosa Penicillin-Binding Protein 3 (PBP3) and N. gonorrhoeae PBP2: Toward a Better Understanding of beta-Lactam Target-Mediated Resistance.
J.Mol.Biol., 431, 2019
|
|
6I1H
 
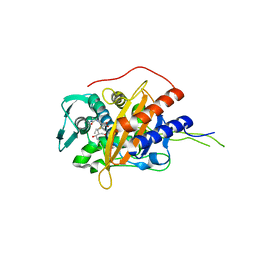 | | Crystal structure of TP domain from Chlamydia trachomatis Penicillin-Binding Protein 3 in complex with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein,Penicillin-binding protein | | Authors: | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | Deposit date: | 2018-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of TP domain from Chlamydia trachomatis Penicillin-Binding Protein 3 in complex with meropenem
To Be Published
|
|
6HZO
 
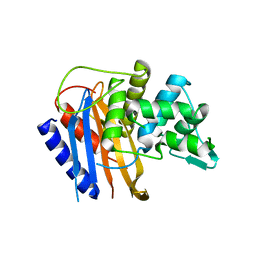 | | Apo structure of TP domain from Haemophilus influenzae Penicillin-Binding Protein 3 | | Descriptor: | FtsI | | Authors: | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | Deposit date: | 2018-10-23 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Novel and Improved Crystal Structures of H. influenzae, E. coli and P. aeruginosa Penicillin-Binding Protein 3 (PBP3) and N. gonorrhoeae PBP2: Toward a Better Understanding of beta-Lactam Target-Mediated Resistance.
J.Mol.Biol., 431, 2019
|
|
6HZQ
 
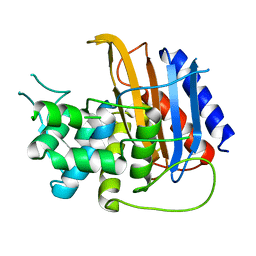 | | Apo structure of TP domain from Escherichia coli Penicillin-Binding Protein 3 | | Descriptor: | Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | Deposit date: | 2018-10-23 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel and Improved Crystal Structures of H. influenzae, E. coli and P. aeruginosa Penicillin-Binding Protein 3 (PBP3) and N. gonorrhoeae PBP2: Toward a Better Understanding of beta-Lactam Target-Mediated Resistance.
J.Mol.Biol., 431, 2019
|
|
6HZH
 
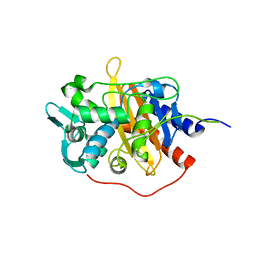 | |
6HZI
 
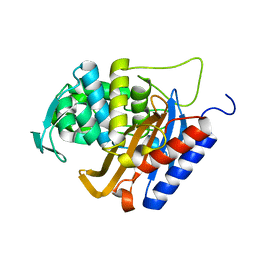 | |
6I1I
 
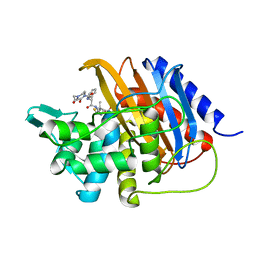 | | Crystal structure of TP domain from Escherichia coli penicillin-binding protein 3 in complex with penicillin | | Descriptor: | Peptidoglycan D,D-transpeptidase FtsI,Peptidoglycan D,D-transpeptidase FtsI, Piperacillin (Open Form) | | Authors: | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | Deposit date: | 2018-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Novel and Improved Crystal Structures of H. influenzae, E. coli and P. aeruginosa Penicillin-Binding Protein 3 (PBP3) and N. gonorrhoeae PBP2: Toward a Better Understanding of beta-Lactam Target-Mediated Resistance.
J.Mol.Biol., 431, 2019
|
|
6I1F
 
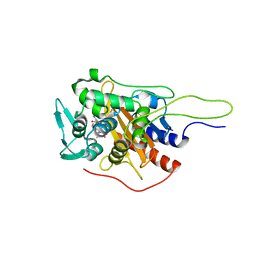 | | Crystal structure of TP domain from Chlamydia trachomatis Penicillin-Binding Protein 3 in complex with amoxicillin | | Descriptor: | 2-{1-[2-AMINO-2-(4-HYDROXY-PHENYL)-ACETYLAMINO]-2-OXO-ETHYL}-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, Penicillin-binding protein,Penicillin-binding protein | | Authors: | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | Deposit date: | 2018-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of TP domain from Chlamydia trachomatis Penicillin-Binding Protein 3 in complex with amoxicillin
To Be Published
|
|
6I1G
 
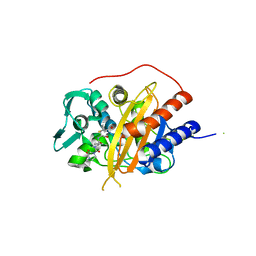 | |
7PSP
 
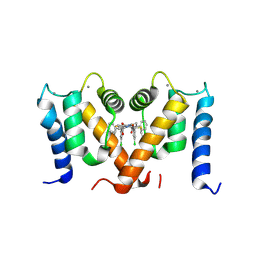 | | Crystal structure of S100A4 labeled with NU000846b. | | Descriptor: | (2R,4R)-1-(2-chloranylethanoyl)-N-(3-chlorophenyl)-4-phenyl-pyrrolidine-2-carboxamide, CALCIUM ION, Protein S100-A4 | | Authors: | Giroud, C, Szommer, T, Coxon, C, Monteiro, O, Christott, T, Bennett, J, Aitmakhanova, K, Raux, B, Newman, J, Elkins, J, Arruda Bezerra, G, Krojer, T, Koekemoer, L, Von Delft, F, Bountr, C, Brennan, P, Fedorov, O. | | Deposit date: | 2021-09-23 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of S100A4 labeled with NU000846b.
To Be Published
|
|
8POA
 
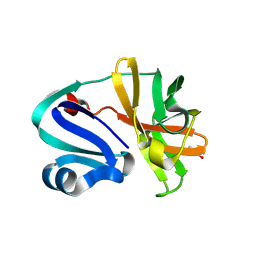 | | Structure of Coxsackievirus A16 (G-10) 2A protease | | Descriptor: | GLYCEROL, Protease 2A, ZINC ION | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-07-04 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of EV A71 2A protease - to be published
To Be Published
|
|
