1UAQ
 
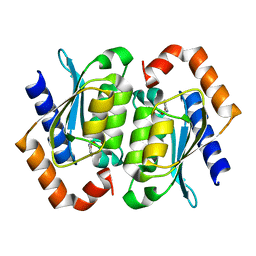 | | The crystal structure of yeast cytosine deaminase | | Descriptor: | DIHYDROPYRIMIDINE-2,4(1H,3H)-DIONE, ZINC ION, cytosine deaminase | | Authors: | Ko, T.-P, Lin, J.-J, Hu, C.-Y, Hsu, Y.-H, Wang, A.H.-J, Liaw, S.-H. | | Deposit date: | 2003-03-14 | | Release date: | 2003-04-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of yeast cytosine deaminase. Insights into enzyme mechanism and evolution
J.Biol.Chem., 278, 2003
|
|
1WD0
 
 | | Crystal structures of the hyperthermophilic chromosomal protein Sac7d in complex with DNA decamers | | Descriptor: | 5'-D(*CP*CP*TP*AP*TP*AP*TP*AP*GP*G)-3', DNA-binding proteins 7a/7b/7d | | Authors: | Ko, T.-P, Chu, H.-M, Chen, C.-Y, Chou, C.-C, Wang, A.H.-J. | | Deposit date: | 2004-05-10 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the hyperthermophilic chromosomal protein Sac7d in complex with DNA decamers.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1WD1
 
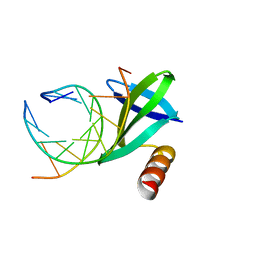 | | Crystal structures of the hyperthermophilic chromosomal protein Sac7d in complex with DNA decamers | | Descriptor: | 5'-D(*CP*CP*TP*AP*CP*GP*TP*AP*GP*G)-3', DNA-binding proteins 7a/7b/7d | | Authors: | Ko, T.-P, Chu, H.-M, Chen, C.-Y, Chou, C.-C, Wang, A.H.-J. | | Deposit date: | 2004-05-10 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the hyperthermophilic chromosomal protein Sac7d in complex with DNA decamers.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1UNK
 
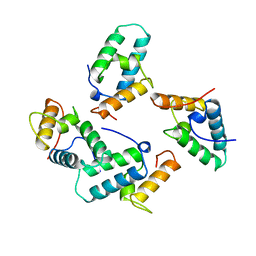 | | STRUCTURE OF COLICIN E7 IMMUNITY PROTEIN | | Descriptor: | COLICIN E7 | | Authors: | Ko, T.-P, Hsieh, S.-Y, Ku, W.-Y, Tseng, M.-Y, Chak, K.-F, Yuan, H.S. | | Deposit date: | 1996-06-21 | | Release date: | 1998-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel role of ImmE7 in the autoregulatory expression of the ColE7 operon and identification of possible RNase active sites in the crystal structure of dimeric ImmE7.
EMBO J., 16, 1997
|
|
1THV
 
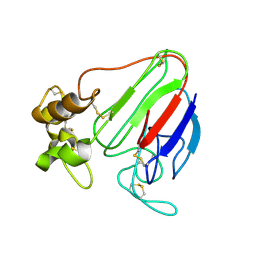 | | THE STRUCTURES OF THREE CRYSTAL FORMS OF THE SWEET PROTEIN THAUMATIN | | Descriptor: | THAUMATIN ISOFORM A | | Authors: | Ko, T.-P, Day, J, Greenwood, A, McPherson, A. | | Deposit date: | 1994-06-10 | | Release date: | 1994-12-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of three crystal forms of the sweet protein thaumatin.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1THW
 
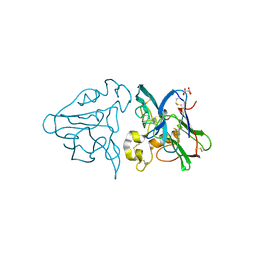 | | THE STRUCTURES OF THREE CRYSTAL FORMS OF THE SWEET PROTEIN THAUMATIN | | Descriptor: | L(+)-TARTARIC ACID, THAUMATIN | | Authors: | Ko, T.-P, Day, J, Greenwood, A, McPherson, A. | | Deposit date: | 1994-06-10 | | Release date: | 1994-12-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of three crystal forms of the sweet protein thaumatin.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
5Z6O
 
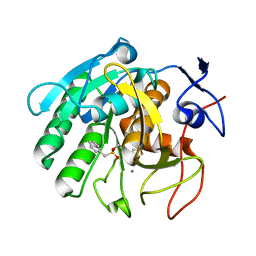 | | Crystal structure of Penicillium cyclopium protease | | Descriptor: | CALCIUM ION, phenylmethanesulfonic acid, protease | | Authors: | Ko, T.-P, Koszelak, S, Ng, J, Day, J, Greenwood, A, McPherson, A. | | Deposit date: | 2018-01-24 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystallographic structure of the subtilisin protease from Penicillium cyclopium.
Biochemistry, 36, 1997
|
|
6ACS
 
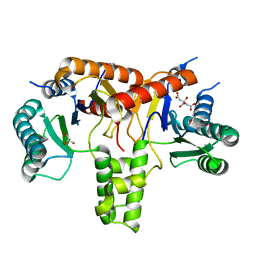 | | poly-cis-prenyltransferase | | Descriptor: | CITRIC ACID, Ditrans,polycis-undecaprenyl-diphosphate synthase ((2E,6E)-farnesyl-diphosphate specific), GLYCEROL, ... | | Authors: | Ko, T.-P, Chen, Y. | | Deposit date: | 2018-07-27 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure of undecaprenyl pyrophosphate synthase from Acinetobacter baumannii
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7EBL
 
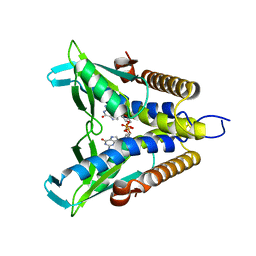 | | Bacterial STING in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), STING | | Authors: | Ko, T.-P, Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2021-03-10 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure and functional implication of bacterial STING.
Nat Commun, 13, 2022
|
|
7EBD
 
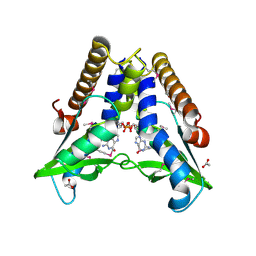 | | Bacterial STING in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ACETATE ION, TIR-like domain-containing protein | | Authors: | Ko, T.-P, Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-15 | | Last modified: | 2022-01-19 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure and functional implication of bacterial STING.
Nat Commun, 13, 2022
|
|
7YIB
 
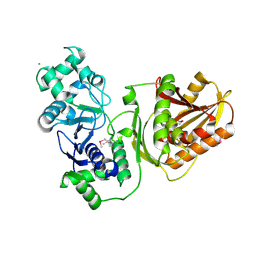 | | Crystal structure of wild-type Cap4 SAVED domain-containing receptor from Enterobacter cloacae | | Descriptor: | CD-NTase-associated protein 4, CHLORIDE ION | | Authors: | Ko, T.-P, Yang, C.-S, Hou, M.-H, Wang, Y.-C, Chen, Y. | | Deposit date: | 2022-07-15 | | Release date: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Specific recognition of cyclic oligonucleotides by Cap4 for phage infection.
Int.J.Biol.Macromol., 237, 2023
|
|
7YIA
 
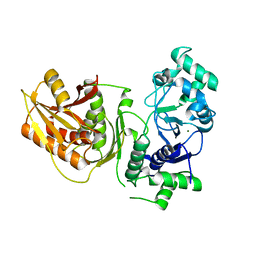 | | Crystal structure of K74A mutant of Cap4 SAVED domain-containing receptor from Enterobacter cloacae | | Descriptor: | CD-NTase-associated protein 4, MAGNESIUM ION | | Authors: | Ko, T.-P, Yang, C.-S, Hou, M.-H, Wang, Y.-C, Chen, Y. | | Deposit date: | 2022-07-15 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.428 Å) | | Cite: | Specific recognition of cyclic oligonucleotides by Cap4 for phage infection.
Int.J.Biol.Macromol., 237, 2023
|
|
4CXP
 
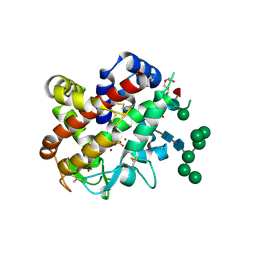 | | Structure of bifunctional endonuclease (AtBFN2) from Arabidopsis thaliana in complex with sulfate | | Descriptor: | ENDONUCLEASE 2, PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Yu, T.-F, Maestre-Reyna, M, Ko, C.-Y, Ko, T.-P, Sun, Y.-J, Lin, T.-Y, Shaw, J.-F, Wang, A.H.-J. | | Deposit date: | 2014-04-08 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Structural Insights of the Ssdna Binding Site in the Multifunctional Endonuclease Atbfn2 from Arabidopsis Thaliana.
Plos One, 9, 2014
|
|
4CWM
 
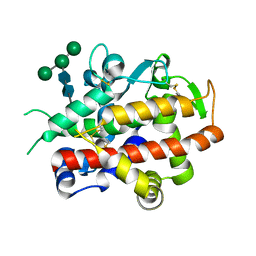 | | High-glycosylation crystal structure of the bifunctional endonuclease (AtBFN2) from Arabidopsis thaliana | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENDONUCLEASE 2, ZINC ION, ... | | Authors: | Yu, T.-F, Maestre-Reyna, M, Ko, C.-Y, Ko, T.-P, Sun, Y.-J, Lin, T.-Y, Shaw, J.-F, Wang, A.H.-J. | | Deposit date: | 2014-04-03 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Insights of the Ssdna Binding Site in the Multifunctional Endonuclease Atbfn2 from Arabidopsis Thaliana.
Plos One, 9, 2014
|
|
4CXO
 
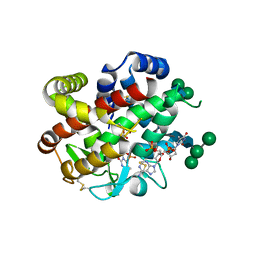 | | bifunctional endonuclease in complex with ssDNA | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, ENDONUCLEASE 2, SULFATE ION, ... | | Authors: | Yu, T.-F, Maestre-Reyna, M, Ko, C.-Y, Ko, T.-P, Sun, Y.-J, Lin, T.-Y, Shaw, J.-F, Wang, A.H.-J. | | Deposit date: | 2014-04-08 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural Insights of the Ssdna Binding Site in the Multifunctional Endonuclease Atbfn2 from Arabidopsis Thaliana.
Plos One, 9, 2014
|
|
4CXV
 
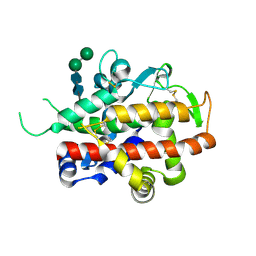 | | Structure of bifunctional endonuclease (AtBFN2) in complex with phosphate. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENDONUCLEASE 2, ... | | Authors: | Yu, T.-F, Maestre-Reyna, M, Ko, C.-Y, Ko, T.-P, Sun, Y.-J, Lin, T.-Y, Shaw, J.-F, Wang, A.H.-J. | | Deposit date: | 2014-04-09 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights of the Ssdna Binding Site in the Multifunctional Endonuclease Atbfn2 from Arabidopsis Thaliana.
Plos One, 9, 2014
|
|
1M7J
 
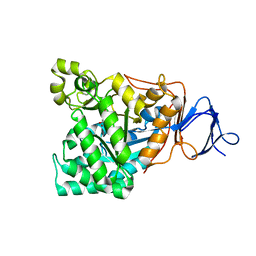 | | Crystal structure of D-aminoacylase defines a novel subset of amidohydrolases | | Descriptor: | ACETATE ION, D-aminoacylase, ZINC ION | | Authors: | Liaw, S.-H, Chen, S.-J, Ko, T.-P, Hsu, C.-S, Wang, A.H.-J, Tsai, Y.-C. | | Deposit date: | 2002-07-22 | | Release date: | 2003-02-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of D-Aminoacylase from Alcaligenes faecalis DA1. A NOVEL SUBSET OF AMIDOHYDROLASES AND INSIGHTS INTO THE ENZYME MECHANISM.
J.Biol.Chem., 278, 2003
|
|
3WG9
 
 | | Crystal structure of RSP, a Rex-family repressor | | Descriptor: | Redox-sensing transcriptional repressor rex, SULFATE ION | | Authors: | Zheng, Y, Ko, T.-P, Guo, R.-T. | | Deposit date: | 2013-08-03 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Distinct structural features of Rex-family repressors to sense redox levels in anaerobes and aerobes.
J.Struct.Biol., 188, 2014
|
|
3FWN
 
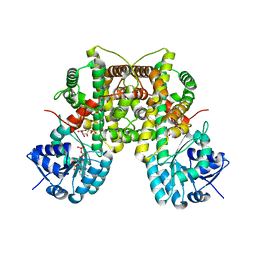 | | Dimeric 6-phosphogluconate dehydrogenase complexed with 6-phosphogluconate and 2'-monophosphoadenosine-5'-diphosphate | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, 6-PHOSPHOGLUCONIC ACID, 6-phosphogluconate dehydrogenase, ... | | Authors: | Chen, Y.-Y, Ko, T.-P, Lo, L.-P, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2009-01-19 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational changes associated with cofactor/substrate binding of 6-phosphogluconate dehydrogenase from Escherichia coli and Klebsiella pneumoniae: Implications for enzyme mechanism
J.Struct.Biol., 169, 2010
|
|
4EM2
 
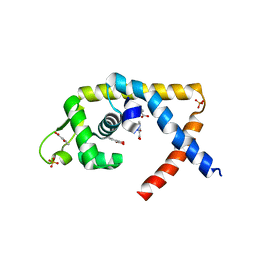 | |
7V6H
 
 | | Crystal Structure of the SpnL | | Descriptor: | Cyclopropane fatty-acyl-phospholipid synthase-like methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wu, H.-H, Ko, T.-P, Liu, H.-W, Tsai, M.-D. | | Deposit date: | 2021-08-20 | | Release date: | 2021-12-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.054 Å) | | Cite: | Evidence for an Enzyme-Catalyzed Rauhut-Currier Reaction during the Biosynthesis of Spinosyn A.
J.Am.Chem.Soc., 143, 2021
|
|
2D45
 
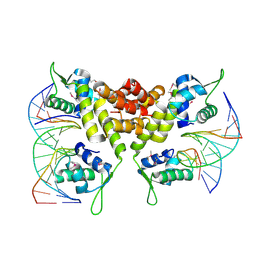 | | Crystal structure of the MecI-mecA repressor-operator complex | | Descriptor: | 5'-D(P*TP*AP*CP*TP*AP*CP*AP*TP*AP*TP*GP*TP*AP*GP*TP*A)-3', Methicillin resistance regulatory protein mecI | | Authors: | Safo, M.K, Ko, T.-P, Musayev, F.N, Zhao, Q, Wang, A.H.-J, Archer, G.L. | | Deposit date: | 2005-10-09 | | Release date: | 2005-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the MecI repressor from Staphylococcus aureus in complex with the cognate DNA operator of mec.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
3PID
 
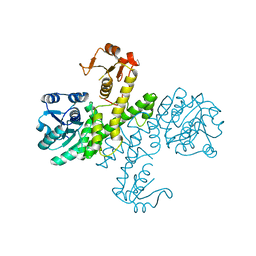 | | The apo-form UDP-glucose 6-dehydrogenase with a C-terminal six-histidine tag | | Descriptor: | UDP-glucose 6-dehydrogenase | | Authors: | Chen, Y.-Y, Ko, T.-P, Lin, C.-H, Chen, W.-H, Wang, A.H.-J. | | Deposit date: | 2010-11-06 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational change upon product binding to Klebsiella pneumoniae UDP-glucose dehydrogenase: a possible inhibition mechanism for the key enzyme in polymyxin resistance.
J.Struct.Biol., 175, 2011
|
|
3PJG
 
 | | Crystal structure of UDP-glucose dehydrogenase from Klebsiella pneumoniae complexed with product UDP-glucuronic acid | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, UDP-glucose 6-dehydrogenase, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Chen, Y.-Y, Ko, T.-P, Lin, C.-H, Chen, W.-H, Wang, A.H.-J. | | Deposit date: | 2010-11-10 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational change upon product binding to Klebsiella pneumoniae UDP-glucose dehydrogenase: a possible inhibition mechanism for the key enzyme in polymyxin resistance.
J.Struct.Biol., 175, 2011
|
|
3PLR
 
 | | Crystal structure of Klebsiella pneumoniae UDP-glucose 6-dehydrogenase complexed with NADH and UDP-glucose | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, UDP-glucose 6-dehydrogenase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Chen, Y.-Y, Ko, T.-P, Lin, C.-H, Chen, W.-H, Wang, A.H.-J. | | Deposit date: | 2010-11-15 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational change upon product binding to Klebsiella pneumoniae UDP-glucose dehydrogenase: a possible inhibition mechanism for the key enzyme in polymyxin resistance.
J.Struct.Biol., 175, 2011
|
|
