3M1K
 
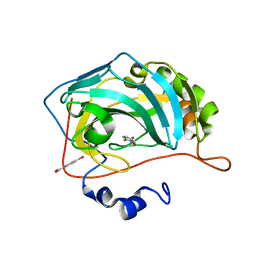 | | Carbonic Anhydrase in complex with fragment | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 1-hydroxy-2-sulfanylpyridinium, Carbonic anhydrase 2, ... | | Authors: | Schulze Wischeler, J, Heine, A, Klebe, G. | | Deposit date: | 2010-03-05 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Bidentate Zinc chelators for alpha-carbonic anhydrases that produce a trigonal bipyramidal coordination geometry.
Chemmedchem, 5, 2010
|
|
3M04
 
 | | Carbonic Anhydrase II in complex with novel sulfonamide inhibitor | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, Carbonic anhydrase 2, ZINC ION, ... | | Authors: | Schulze Wischeler, J, Heine, A, Klebe, G, Sandner, N.U. | | Deposit date: | 2010-03-02 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural investigation and inhibitor studies on Carbonic Anhydrase II
To be Published
|
|
3M0I
 
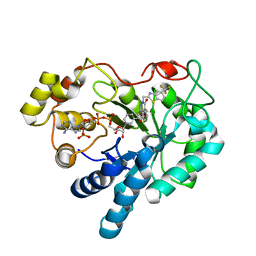 | | Human Aldose Reductase mutant T113V in complex with Zopolrestat | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldose Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-03-03 | | Release date: | 2011-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Ligand-induced fit affects binding modes and provokes changes in crystal packing of aldose reductase
Biochim.Biophys.Acta, 1810, 2011
|
|
3M5S
 
 | |
3M1Q
 
 | |
3M2Z
 
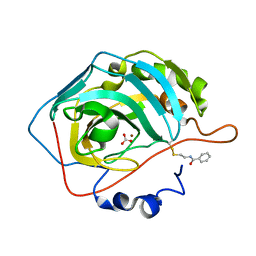 | |
3M4H
 
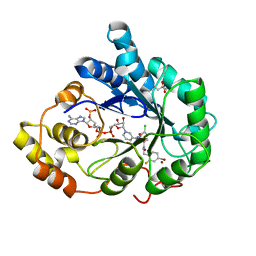 | | Human Aldose Reductase mutant T113V complexed with IDD388 | | Descriptor: | (2-{[(4-BROMO-2-FLUOROBENZYL)AMINO]CARBONYL}-5-CHLOROPHENOXY)ACETIC ACID, Aldose reductase, BROMIDE ION, ... | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-03-11 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Tracing the detail: how mutations affect binding modes and thermodynamic signatures of closely related aldose reductase inhibitors
J.Mol.Biol., 406, 2011
|
|
3M2X
 
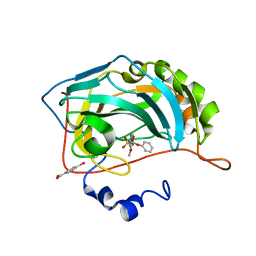 | |
3M64
 
 | | Human aldose reductase mutant T113V complexed with IDD393 | | Descriptor: | (5-CHLORO-2-{[(3-NITROBENZYL)AMINO]CARBONYL}PHENOXY)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-03-15 | | Release date: | 2011-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Ligand-induced fit affects binding modes and provokes changes in crystal packing of aldose reductase
Biochim.Biophys.Acta, 1810, 2011
|
|
3M5T
 
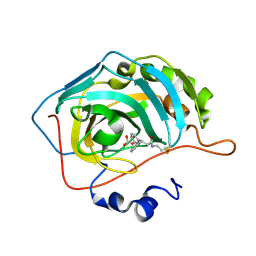 | |
3ONB
 
 | | Bond breakage and relocation of a covalently bound bromine of IDD594 in a complex with hAR T113A mutant after extensive radiation dose | | Descriptor: | Aldose reductase, BROMIDE ION, CITRIC ACID, ... | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Radiation damage reveals promising interaction position
J.SYNCHROTRON RADIAT., 18, 2011
|
|
3PB5
 
 | | Endothiapepsin in complex with a fragment | | Descriptor: | Endothiapepsin, GLYCEROL, N-methyl-1-[5-(pyridin-3-yloxy)furan-2-yl]methanamine | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-10-20 | | Release date: | 2011-10-19 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A small nonrule of 3 compatible fragment library provides high hit rate of endothiapepsin crystal structures with various fragment chemotypes.
J.Med.Chem., 54, 2011
|
|
3P2V
 
 | | Novel Benzothiazepine Inhibitor in Complex with human Aldose Reductase | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [(2S)-4-oxo-2-phenyl-3,4-dihydro-1,5-benzothiazepin-5(2H)-yl]acetic acid | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-10-04 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Ligand-induced fit affects binding modes and provokes changes in crystal packing of aldose reductase
Biochim.Biophys.Acta, 1810, 2011
|
|
3LQG
 
 | | Human aldose reductase mutant T113A complexed with IDD388 | | Descriptor: | (2-{[(4-BROMO-2-FLUOROBENZYL)AMINO]CARBONYL}-5-CHLOROPHENOXY)ACETIC ACID, Aldose reductase, BROMIDE ION, ... | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-02-09 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Tracing the detail: how mutations affect binding modes and thermodynamic signatures of closely related aldose reductase inhibitors
J.Mol.Biol., 406, 2011
|
|
3LEP
 
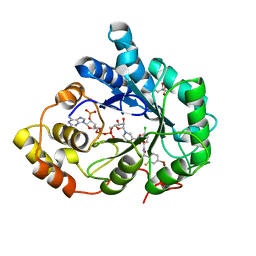 | | Human Aldose Reductase mutant T113C in complex with IDD388 | | Descriptor: | (2-{[(4-BROMO-2-FLUOROBENZYL)AMINO]CARBONYL}-5-CHLOROPHENOXY)ACETIC ACID, Aldose reductase, BROMIDE ION, ... | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-01-15 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Tracing the detail: how mutations affect binding modes and thermodynamic signatures of closely related aldose reductase inhibitors
J.Mol.Biol., 406, 2011
|
|
3LJJ
 
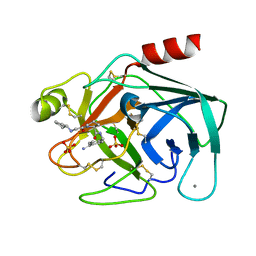 | | Bovine trypsin in complex with UB-THR 10 | | Descriptor: | (S)-N-(4-carbamimidoylbenzyl)-1-(2-(cyclopentylamino)ethanoyl)pyrrolidine-2-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Wegscheid-Gerlach, C, Heine, A, Klebe, G. | | Deposit date: | 2010-01-26 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Congeneric but still distinct: how closely related trypsin ligands exhibit different thermodynamic and structural properties.
J.Mol.Biol., 405, 2011
|
|
3ONC
 
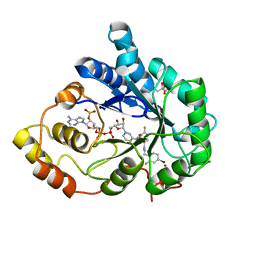 | | Bond breakage and relocation of a covalently bound bromine of IDD594 in a complex with hAR T113A mutant after moderate radiation dose | | Descriptor: | Aldose reductase, BROMIDE ION, CITRIC ACID, ... | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Radiation damage reveals promising interaction position
J.SYNCHROTRON RADIAT., 18, 2011
|
|
3PBD
 
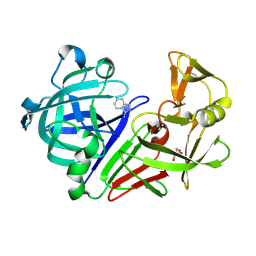 | | Endothiapepsin in complex with a fragment | | Descriptor: | 1H-isoindol-3-amine, Endothiapepsin, GLYCEROL | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-10-20 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A small nonrule of 3 compatible fragment library provides high hit rate of endothiapepsin crystal structures with various fragment chemotypes.
J.Med.Chem., 54, 2011
|
|
3PBZ
 
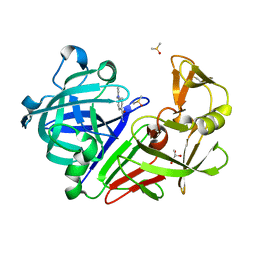 | | Endothiapepsin in complex with a fragment | | Descriptor: | 4-(diethylamino)benzohydrazide, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-10-21 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | A small nonrule of 3 compatible fragment library provides high hit rate of endothiapepsin crystal structures with various fragment chemotypes.
J.Med.Chem., 54, 2011
|
|
3PCW
 
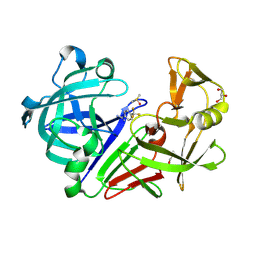 | | Endothiapepsin in complex with a fragment | | Descriptor: | 4-(trifluoromethyl)benzenecarboximidamide, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-10-22 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A small nonrule of 3 compatible fragment library provides high hit rate of endothiapepsin crystal structures with various fragment chemotypes.
J.Med.Chem., 54, 2011
|
|
3PM4
 
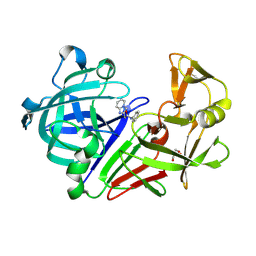 | | Endothiapepsin in complex with a fragment | | Descriptor: | 2-(imidazo[1,2-a]pyridin-2-yl)-N-phenylacetamide, Endothiapepsin, GLYCEROL | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-11-16 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A small nonrule of 3 compatible fragment library provides high hit rate of endothiapepsin crystal structures with various fragment chemotypes.
J.Med.Chem., 54, 2011
|
|
3PGI
 
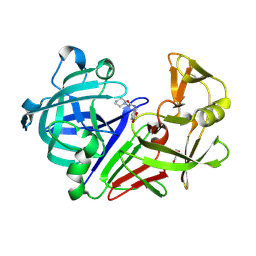 | | Endothiapepsin in complex with a fragment | | Descriptor: | Endothiapepsin, GLYCEROL, N-(1,3-benzodioxol-5-yl)-2-(piperidin-1-yl)acetamide | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-11-02 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A small nonrule of 3 compatible fragment library provides high hit rate of endothiapepsin crystal structures with various fragment chemotypes.
J.Med.Chem., 54, 2011
|
|
3PI0
 
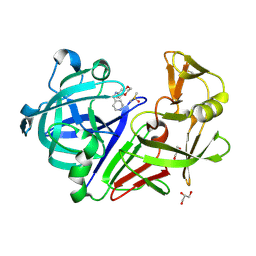 | | Endothiapepsin in complex with a fragment | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin, GLYCEROL, ... | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-11-05 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A small nonrule of 3 compatible fragment library provides high hit rate of endothiapepsin crystal structures with various fragment chemotypes.
J.Med.Chem., 54, 2011
|
|
3POO
 
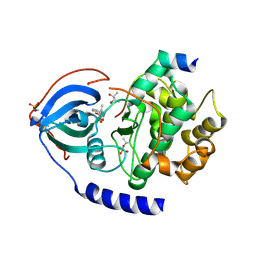 | | human cAMP-dependent protein kinase in complex with an inhibitor | | Descriptor: | (2S,3S)-butane-2,3-diol, N'-[(E)-(2,4-dihydroxy-6-methylphenyl)methylidene]-2-(3-methoxyphenyl)acetohydrazide, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-11-23 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | fragment based drug design on PKA
To be Published
|
|
3Q70
 
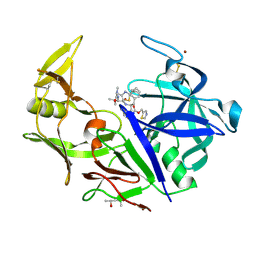 | |
