6LZ9
 
 | |
1V4N
 
 | | Structure of 5'-deoxy-5'-methylthioadenosine phosphorylase homologue from Sulfolobus tokodaii | | Descriptor: | 271aa long hypothetical 5'-methylthioadenosine phosphorylase | | Authors: | Kitago, Y, Yasutake, Y, Sakai, N, Tsujimura, M, Yao, M, Watanabe, N, Kawarabayasi, Y, Tanaka, I. | | Deposit date: | 2003-11-14 | | Release date: | 2005-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of Sulfolobus tokodaii MTAP
To be Published
|
|
5GIS
 
 | | Crystal structure of a Fab fragment with its ligand peptide | | Descriptor: | Heavy chain of Fab fragment, Light chain of Fab fragment, SULFATE ION, ... | | Authors: | Kitago, Y, Kaneko, K.K, Ogasawara, S, Kato, Y, Takagi, J. | | Deposit date: | 2016-06-24 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis for multi-specific peptide recognition by the anti-IDH1/2 monoclonal antibody, MsMab-1.
Biochem. Biophys. Res. Commun., 478, 2016
|
|
5GIR
 
 | | Crystal structure of a Fab fragment with its ligand peptide | | Descriptor: | 1,2-ETHANEDIOL, Heavy chain of Fab fragment, LYS-PRO-ILE-ILE-ILE-GLY-SER-HIS-ALA-TYR-GLY-ASP, ... | | Authors: | Kitago, Y, Kaneko, K.K, Ogasawara, S, Kato, Y, Takagi, J. | | Deposit date: | 2016-06-24 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis for multi-specific peptide recognition by the anti-IDH1/2 monoclonal antibody, MsMab-1.
Biochem. Biophys. Res. Commun., 478, 2016
|
|
5AUM
 
 | |
3WSY
 
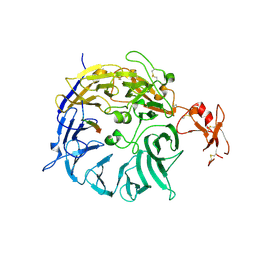 | | SorLA Vps10p domain in complex with its own propeptide fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Sortilin-related receptor, peptide from Sortilin-related receptor | | Authors: | Kitago, Y, Nakata, Z, Nagae, M, Nogi, T, Takagi, J. | | Deposit date: | 2014-03-30 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis for amyloidogenic peptide recognition by sorLA.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3WSZ
 
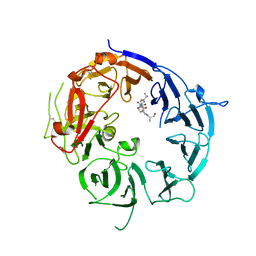 | | SorLA Vps10p domain in complex with Abeta-derived peptide | | Descriptor: | 10-mer peptide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Sortilin-related receptor | | Authors: | Kitago, Y, Nakata, Z, Nagae, M, Nogi, T, Takagi, J. | | Deposit date: | 2014-03-30 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Structural basis for amyloidogenic peptide recognition by sorLA.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3WSX
 
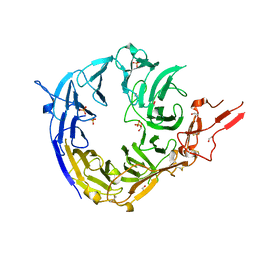 | | SorLA Vps10p domain in ligand-free form | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Kitago, Y, Nakata, Z, Nagae, M, Nogi, T, Takagi, J. | | Deposit date: | 2014-03-30 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for amyloidogenic peptide recognition by sorLA.
Nat.Struct.Mol.Biol., 22, 2015
|
|
2E6U
 
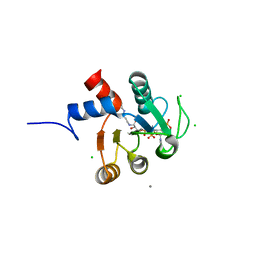 | | Crystal structure of hypothetical protein PH1109 from Pyrococcus horikoshii | | Descriptor: | CALCIUM ION, CHLORIDE ION, COENZYME A, ... | | Authors: | Kitago, Y, Min, Y, Watanabe, N, Tanaka, I. | | Deposit date: | 2007-01-03 | | Release date: | 2007-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure determination of a novel protein by sulfur SAD using chromium radiation in combination with a new crystal-mounting method
ACTA CRYSTALLOGR.,SECT.D, 61, 2005
|
|
2EJ1
 
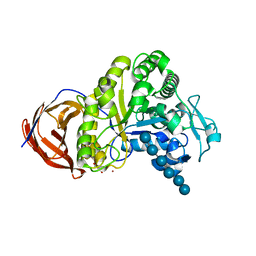 | | Crystal structure of Cel44A, GH family 44 endoglucanase from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CHLORIDE ION, Endoglucanase, ... | | Authors: | Kitago, Y, Karita, S, Watanabe, N, Sakka, K, Tanaka, I. | | Deposit date: | 2007-03-14 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Cel44A, a glycoside hydrolase family 44 endoglucanase from Clostridium thermocellum.
J.Biol.Chem., 282, 2007
|
|
2E0P
 
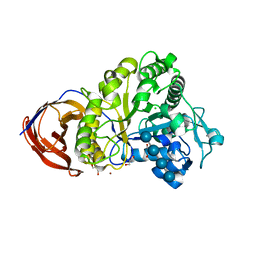 | | The crystal structure of Cel44A | | Descriptor: | CALCIUM ION, CHLORIDE ION, Endoglucanase, ... | | Authors: | Kitago, Y, Karita, S, Watanabe, N, Sakka, K, Tanaka, I. | | Deposit date: | 2006-10-11 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Cel44A, a glycoside hydrolase family 44 endoglucanase from Clostridium thermocellum.
J.Biol.Chem., 282, 2007
|
|
2EQD
 
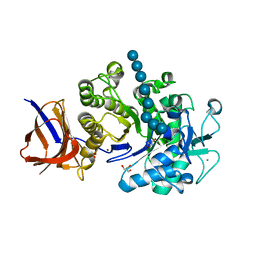 | | Crystal structure of Cel44A, GH family 44 endoglucanase from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CHLORIDE ION, Endoglucanase, ... | | Authors: | Kitago, Y, Karita, S, Watanabe, N, Sakka, K, Tanaka, I. | | Deposit date: | 2007-03-30 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Cel44A, a glycoside hydrolase family 44 endoglucanase from Clostridium thermocellum.
J.Biol.Chem., 282, 2007
|
|
2E4T
 
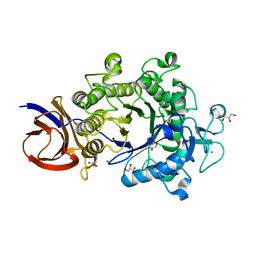 | | Crystal structure of Cel44A, GH family 44 endoglucanase from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CHLORIDE ION, Endoglucanase, ... | | Authors: | Kitago, Y, Karita, S, Watanabe, N, Sakka, K, Tanaka, I. | | Deposit date: | 2006-12-16 | | Release date: | 2007-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Crystal structure of Cel44A, a glycoside hydrolase family 44 endoglucanase from Clostridium thermocellum.
J.Biol.Chem., 282, 2007
|
|
2EEX
 
 | | Crystal structure of Cel44A, GH family 44 endoglucanase from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CHLORIDE ION, Endoglucanase, ... | | Authors: | Kitago, Y, Karita, S, Watanabe, N, Sakka, K, Tanaka, I. | | Deposit date: | 2007-02-19 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Cel44A, a glycoside hydrolase family 44 endoglucanase from Clostridium thermocellum.
J.Biol.Chem., 282, 2007
|
|
2CVI
 
 | | Crystal structure of hypothetical protein PHS023 from Pyrococcus horikoshii | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 75aa long hypothetical regulatory protein AsnC, POTASSIUM ION | | Authors: | Kitago, Y, Sakai, N, Matsumoto, D, Yao, M, Aizawa, T, Demura, M, Kawano, K, Nitta, K, Watanabe, N, Tanaka, I. | | Deposit date: | 2005-06-03 | | Release date: | 2006-06-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of hypothetical protein PHS023 from Pyrococcus horikoshii
TO BE PUBLISHED
|
|
4YO0
 
 | | Crystal structure of monoclonal anti-human podoplanin antibody NZ-1 with bound PA peptide | | Descriptor: | 1,2-ETHANEDIOL, Heavy chain of antigen binding fragment, Fab, ... | | Authors: | Fujii, Y, Kitago, Y, Arimori, T, Takagi, J. | | Deposit date: | 2015-03-11 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Tailored placement of a turn-forming PA tag into the structured domain of a protein to probe its conformational state
J.Cell.Sci., 129, 2016
|
|
4YNY
 
 | | Crystal structure of monoclonal anti-human podoplanin antibody NZ-1 | | Descriptor: | Heavy chain of antigen binding fragment, Fab, Light chain of antigen binding fragment | | Authors: | Fujii, Y, Kitago, Y, Arimori, T, Takagi, J. | | Deposit date: | 2015-03-11 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.584 Å) | | Cite: | Tailored placement of a turn-forming PA tag into the structured domain of a protein to probe its conformational state
J.Cell.Sci., 129, 2016
|
|
5B4W
 
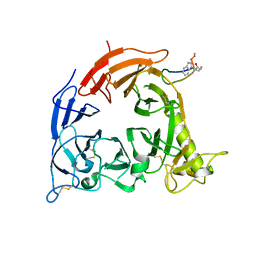 | | Crystal structure of Plexin inhibitor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-B1, Synthesized cyclic peptide | | Authors: | Matsunaga, Y, Kitago, Y, Arimori, T, Takagi, J. | | Deposit date: | 2016-04-19 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Allosteric Inhibition of a Semaphorin 4D Receptor Plexin B1 by a High-Affinity Macrocyclic Peptide
Cell Chem Biol, 23, 2016
|
|
2ZY6
 
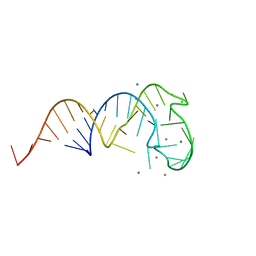 | | Crystal structure of a truncated tRNA, TPHE39A | | Descriptor: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Tanaka, I, Yao, M, Tanaka, Y, Kitago, Y, Ymagata, S. | | Deposit date: | 2009-01-14 | | Release date: | 2009-06-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Deduced RNA binding mechanism of ThiI based on structural and binding analyses of a minimal RNA ligand
Rna, 15, 2009
|
|
3A8S
 
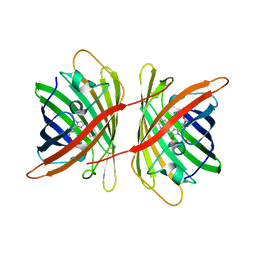 | |
5XAU
 
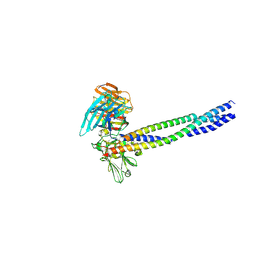 | | Crystal structure of integrin binding fragment of laminin-511 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Takizawa, M, Arimori, T, Kitago, Y, Takagi, J, Sekiguchi, K. | | Deposit date: | 2017-03-15 | | Release date: | 2017-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanistic basis for the recognition of laminin-511 by alpha 6 beta 1 integrin.
Sci Adv, 3, 2017
|
|
3WBQ
 
 | | Crystal structure of carbohydrate recognition domain of Blood Dendritic Cell Antigen-2 (BDCA2) lectin (crystal form-2) | | Descriptor: | C-type lectin domain family 4 member C | | Authors: | Nagae, M, Ikeda, A, Kitago, Y, Matsumoto, N, Yamamoto, K, Yamaguchi, Y. | | Deposit date: | 2013-05-20 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of carbohydrate recognition domain of blood dendritic cell antigen-2 (BDCA2) reveal a common domain-swapped dimer.
Proteins, 82, 2014
|
|
3WBP
 
 | | Crystal structure of carbohydrate recognition domain of Blood Dendritic Cell Antigen-2 (BDCA2) lectin (crystal form-1) | | Descriptor: | 1,2-ETHANEDIOL, C-type lectin domain family 4 member C | | Authors: | Nagae, M, Ikeda, A, Kitago, Y, Matsumoto, N, Yamamoto, K, Yamaguchi, Y. | | Deposit date: | 2013-05-20 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of carbohydrate recognition domain of blood dendritic cell antigen-2 (BDCA2) reveal a common domain-swapped dimer.
Proteins, 82, 2014
|
|
3WBR
 
 | | Crystal structure of carbohydrate recognition domain of Blood Dendritic Cell Antigen-2 (BDCA2) lectin (crystal form-3) | | Descriptor: | C-type lectin domain family 4 member C | | Authors: | Nagae, M, Ikeda, A, Kitago, Y, Matsumoto, N, Yamamoto, K, Yamaguchi, Y. | | Deposit date: | 2013-05-20 | | Release date: | 2013-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of carbohydrate recognition domain of blood dendritic cell antigen-2 (BDCA2) reveal a common domain-swapped dimer.
Proteins, 82, 2014
|
|
