6OK1
 
 | | Ltp2-ChsH2(DUF35) aldolase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ChsH2(DUF35), ... | | Authors: | Kimber, M.S, Mallette, E, Aggett, R, Seah, S.Y.K. | | Deposit date: | 2019-04-12 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The steroid side-chain-cleaving aldolase Ltp2-ChsH2DUF35is a thiolase superfamily member with a radically repurposed active site.
J.Biol.Chem., 294, 2019
|
|
5SWC
 
 | | The structure of the beta-carbonic anhydrase CcaA | | Descriptor: | CHLORIDE ION, Carbonic anhydrase, FORMIC ACID, ... | | Authors: | Kimber, M.S, McGurn, L, White, S.A. | | Deposit date: | 2016-08-08 | | Release date: | 2016-10-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure, kinetics and interactions of the beta-carboxysomal beta-carbonic anhydrase, CcaA.
Biochem. J., 473, 2016
|
|
6MGC
 
 | | Escherichia coli KpsC, N-terminal domain | | Descriptor: | CHLORIDE ION, CYTIDINE-5'-MONOPHOSPHATE, Capsule polysaccharide export protein KpsC, ... | | Authors: | Doyle, L, Mallette, E, Kimber, M.S. | | Deposit date: | 2018-09-13 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Biosynthesis of a conserved glycolipid anchor for Gram-negative bacterial capsules.
Nat.Chem.Biol., 15, 2019
|
|
1HYB
 
 | | CRYSTAL STRUCTURE OF AN ACTIVE SITE MUTANT OF METHANOBACTERIUM THERMOAUTOTROPHICUM NICOTINAMIDE MONONUCLEOTIDE ADENYLYLTRANSFERASE | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NICOTINAMIDE MONONUCLEOTIDE ADENYLYLTRANSFERASE, SULFATE ION | | Authors: | Saridakis, V, Christendat, D, Kimber, M.S, Edwards, A.M, Pai, E.F. | | Deposit date: | 2001-01-18 | | Release date: | 2001-03-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into ligand binding and catalysis of a central step in NAD+ synthesis: structures of Methanobacterium thermoautotrophicum NMN adenylyltransferase complexes.
J.Biol.Chem., 276, 2001
|
|
6EF6
 
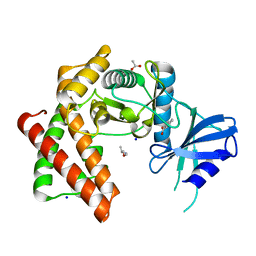 | | Structure of the microcompartment-associated aminopropanol kinase | | Descriptor: | (2R)-1-methoxypropan-2-amine, ACETATE ION, Aminoglycoside phosphotransferase, ... | | Authors: | Mallette, E, Kimber, M.S. | | Deposit date: | 2018-08-16 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and kinetic characterization of (S)-1-amino-2-propanol kinase from the aminoacetone utilization microcompartment ofMycobacterium smegmatis.
J.Biol.Chem., 293, 2018
|
|
4N8X
 
 | | The structure of Nostoc sp. PCC 7120 CcmL | | Descriptor: | Carbon dioxide concentrating mechanism protein, SULFATE ION | | Authors: | Keeling, T.J, Kimber, M.S. | | Deposit date: | 2013-10-18 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Interactions and structural variability of beta-carboxysomal shell protein CcmL.
Photosynth.Res., 121, 2014
|
|
8VWR
 
 | | Structure of steroid hydratase from Comamonas testosteroni | | Descriptor: | Acyl dehydratase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schroeter, K.L, Forrester, T.J.B, Kimber, M.S, Seah, S.Y.K. | | Deposit date: | 2024-02-02 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Shy is a proteobacterial steroid hydratase which catalyzes steroid side chain degradation without requiring a catalytically inert partner domain.
J.Biol.Chem., 300, 2024
|
|
7UTF
 
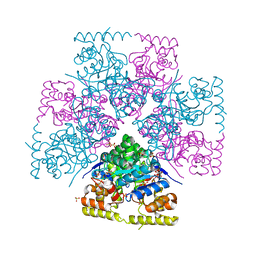 | | Structure-Function characterization of an aldo-keto reductase involved in detoxification of the mycotoxin, deoxynivalenol | | Descriptor: | CITRATE ANION, Putative oxidoreductase, aryl-alcohol dehydrogenase like protein, ... | | Authors: | Abraham, N, Schroeter, K.L, Kimber, M.S, Seah, S.Y.K. | | Deposit date: | 2022-04-26 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-function characterization of an aldo-keto reductase involved in detoxification of the mycotoxin, deoxynivalenol.
Sci Rep, 12, 2022
|
|
3KWD
 
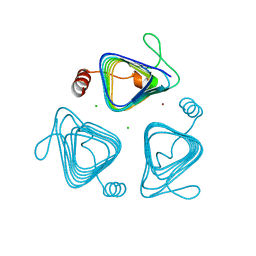 | | Inactive truncation of the beta-carboxysomal gamma-Carbonic Anhydrase, CcmM, form 1 | | Descriptor: | CHLORIDE ION, Carbon dioxide concentrating mechanism protein, ZINC ION | | Authors: | Pena, K.L, Kimber, M.S, Castel, S.E. | | Deposit date: | 2009-12-01 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis of the oxidative activation of the carboxysomal {gamma}-carbonic anhydrase, CcmM.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4JN6
 
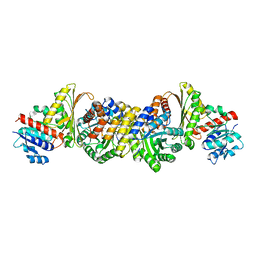 | | Crystal Structure of the Aldolase-Dehydrogenase Complex from Mycobacterium tuberculosis HRv37 | | Descriptor: | 4-hydroxy-2-oxovalerate aldolase, Acetaldehyde dehydrogenase, MANGANESE (II) ION, ... | | Authors: | Carere, J, McKenna, S.E, Kimber, M.S, Seah, S.Y.K. | | Deposit date: | 2013-03-14 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Characterization of an Aldolase-Dehydrogenase Complex from the Cholesterol Degradation Pathway of Mycobacterium tuberculosis.
Biochemistry, 52, 2013
|
|
6CI8
 
 | |
6CI9
 
 | |
8CSE
 
 | | WbbB in complex with alpha-Rha-(1-3)-beta-GlcNAc acceptor | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, N-(8-hydroxyoctyl)-4-methoxybenzamide, N-acetyl glucosaminyl transferase, ... | | Authors: | Forrester, T.J.B, Kimber, M.S. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The retaining beta-Kdo glycosyltransferase WbbB uses a double-displacement mechanism with an intermediate adduct rearrangement step.
Nat Commun, 13, 2022
|
|
8CSD
 
 | | WbbB D232C Kdo adduct | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, CHLORIDE ION, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Forrester, T.J.B, Kimber, M.S. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The retaining beta-Kdo glycosyltransferase WbbB uses a double-displacement mechanism with an intermediate adduct rearrangement step.
Nat Commun, 13, 2022
|
|
8CSB
 
 | | WbbB D232N in complex with CMP-beta-Kdo | | Descriptor: | CYTIDINE 5'-MONOPHOSPHATE 3-DEOXY-BETA-D-GULO-OCT-2-ULO-PYRANOSONIC ACID, CYTIDINE-5'-MONOPHOSPHATE, N-acetyl glucosaminyl transferase, ... | | Authors: | Forrester, T.J.B, Kimber, M.S. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The retaining beta-Kdo glycosyltransferase WbbB uses a double-displacement mechanism with an intermediate adduct rearrangement step.
Nat Commun, 13, 2022
|
|
8CSF
 
 | | WbbB D232C-Kdo adduct + alpha-Rha(1,3)GlcNAc ternary complex | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, CYTIDINE-5'-MONOPHOSPHATE, N-acetyl glucosaminyl transferase, ... | | Authors: | Forrester, T.J.B, Kimber, M.S. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The retaining beta-Kdo glycosyltransferase WbbB uses a double-displacement mechanism with an intermediate adduct rearrangement step.
Nat Commun, 13, 2022
|
|
8CSC
 
 | | WbbB D232N-Kdo adduct | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, CHLORIDE ION, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Forrester, T.J.B, Kimber, M.S. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The retaining beta-Kdo glycosyltransferase WbbB uses a double-displacement mechanism with an intermediate adduct rearrangement step.
Nat Commun, 13, 2022
|
|
5CGZ
 
 | | Crystal structure of GalB, the 4-carboxy-2-hydroxymuconate hydratase, from Pseuodomonas putida KT2440 | | Descriptor: | 4-oxalmesaconate hydratase, GLYCEROL, ZINC ION | | Authors: | Mazurkewich, S, Brott, A.S, Kimber, M.S, Seah, S.Y.K. | | Deposit date: | 2015-07-09 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Structural and Kinetic Characterization of the 4-Carboxy-2-hydroxymuconate Hydratase from the Gallate and Protocatechuate 4,5-Cleavage Pathways of Pseudomonas putida KT2440.
J.Biol.Chem., 291, 2016
|
|
5DKO
 
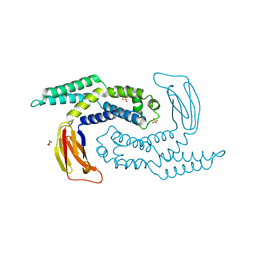 | | The structure of Escherichia coli ZapD | | Descriptor: | Cell division protein ZapD, SULFATE ION | | Authors: | Wroblewski, C, Kimber, M.S. | | Deposit date: | 2015-09-03 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Mutational Analyses of Escherichia coli ZapD Reveal Charged Residues Involved in FtsZ Filament Bundling.
J.Bacteriol., 198, 2016
|
|
5L38
 
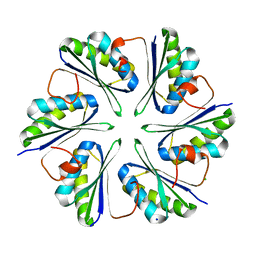 | |
5L37
 
 | |
5L39
 
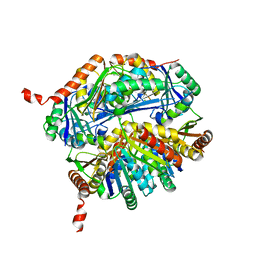 | |
3GGP
 
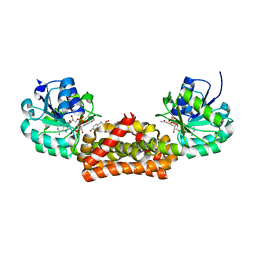 | | Crystal structure of prephenate dehydrogenase from A. aeolicus in complex with hydroxyphenyl propionate and NAD+ | | Descriptor: | CHLORIDE ION, HYDROXYPHENYL PROPIONIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Sun, W, Shahinas, D, Kimber, M.S, Christendat, D. | | Deposit date: | 2009-03-01 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Crystal Structure of Aquifex aeolicus Prephenate Dehydrogenase Reveals the Mode of Tyrosine Inhibition.
J.Biol.Chem., 284, 2009
|
|
3SSQ
 
 | | CcmK2 - form 1 dodecamer | | Descriptor: | CHLORIDE ION, Carbon dioxide concentrating mechanism protein, GLYCEROL | | Authors: | Samborska, B, Kimber, M.S. | | Deposit date: | 2011-07-08 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A CcmK2 double layer is the dominant architectural feature of the beta-carboxysomal shell facet
Structure, 2012
|
|
5FA0
 
 | | The structure of the beta-3-deoxy-D-manno-oct-2-ulosonic acid transferase domain from WbbB | | Descriptor: | CHLORIDE ION, Putative N-acetyl glucosaminyl transferase | | Authors: | Mallette, E, Ovchinnikova, O.G, Whitfield, C, Kimber, M.S. | | Deposit date: | 2015-12-10 | | Release date: | 2016-05-18 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bacterial beta-Kdo glycosyltransferases represent a new glycosyltransferase family (GT99).
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
