3RZP
 
 | | Crystal Structure of the C194A mutant of 7-cyano-7-deazaguanine reductase, QueF from Vibrio cholerae complexed with preQ1 | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, GLYCEROL, NADPH-dependent 7-cyano-7-deazaguanine reductase | | Authors: | Kim, Y, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-12 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the C194A mutant of 7-cyano-7-deazaguanine reductase, QueF from Vibrio cholerae complexed with preQ1
To be Published
|
|
1PV5
 
 | |
1QWR
 
 | |
1RYD
 
 | | Crystal Structure of Glucose-Fructose Oxidoreductase from Zymomonas mobilis | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Kim, Y, Arora, M, Straza, M, Joachimiak, A. | | Deposit date: | 2003-12-22 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Glucose-Fructose Oxidoreductase from Zymomonas mobilis
To be Published
|
|
1S5A
 
 | |
1RLI
 
 | |
1RFZ
 
 | | Structure of Protein of Unknown Function from Bacillus stearothermophilus | | Descriptor: | Hypothetical protein APC35681, SULFATE ION | | Authors: | Kim, Y, Wu, R, Cuff, M.E, Quartey, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-11-10 | | Release date: | 2004-03-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of hypothetical protein APC35681 from Bacillus stearothermophilus
To be Published
|
|
1S5U
 
 | | Crystal Structure of Hypothetical Protein EC709 from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, Protein ybgC, SULFATE ION | | Authors: | Kim, Y, Joachimiak, A, Skarina, T, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-01-21 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Hypothetical Protein EC709 from Escherichia coli
To be Published
|
|
1RYE
 
 | | Crystal Structure of the Shifted Form of the Glucose-Fructose Oxidoreductase from Zymomonas mobilis | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Kim, Y, Arora, M, Straza, M, Donnelly, M, Joachimiak, A. | | Deposit date: | 2003-12-22 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Shifted Form of the Glucose-Fructose Oxidoreductase from Zymomonas mobilis
To be Published
|
|
1U7N
 
 | | Crystal Structure of the fatty acid/phospholipid synthesis protein PlsX from Enterococcus faecalis V583 | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Fatty acid/phospholipid synthesis protein plsX | | Authors: | Kim, Y, Li, H, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-04 | | Release date: | 2004-08-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of fatty acid/phospholipid synthesis protein PlsX from Enterococcus faecalis.
J.STRUCT.FUNCT.GENOM., 10, 2009
|
|
1TU9
 
 | | Crystal Structure of a Protein PA3967, a Structurally Highly Homologous to a Human Hemoglobin, from Pseudomonas aeruginosa PAO1 | | Descriptor: | 1,2-ETHANEDIOL, PROPANOIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kim, Y, Joachimiak, A, Skarina, T, Egorova, O, Bochkarev, A, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-24 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of PA3967 from Pseudomonas aeruginosa PAO1, a Hypothetical Protein which is highly homologous to human Hemoglobin in structure.
To be Published
|
|
1Q9U
 
 | |
1U84
 
 | | Crystal Structure of APC36109 from Bacillus stearothermophilus | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Hypothetical protein | | Authors: | Kim, Y, Zhou, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-04 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of APC36109 from Bacillus stearothermophilus
To be Published
|
|
1TZ0
 
 | |
1T07
 
 | | Crystal Structure of Conserved Protein of Unknown Function PA5148 from Pseudomonas aeruginosa | | Descriptor: | Hypothetical UPF0269 protein PA5148 | | Authors: | Kim, Y, Joachimiak, A, Skarina, T, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-04-07 | | Release date: | 2004-08-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of PA5148 from Pseudomonas aeruginosa
To be Published
|
|
1SS4
 
 | |
1T57
 
 | | Crystal Structure of the Conserved Protein MTH1675 from Methanobacterium thermoautotrophicum | | Descriptor: | Conserved Protein MTH1675, FLAVIN MONONUCLEOTIDE, MAGNESIUM ION | | Authors: | Kim, Y, Joachimiak, A, Saridakis, V, Xu, X, Arrowsmith, C.H, Christendat, D, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-03 | | Release date: | 2004-08-03 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Conserved Protein MTH1675 from Methanobacterium thermoautotrophicum
To be Published
|
|
1TAQ
 
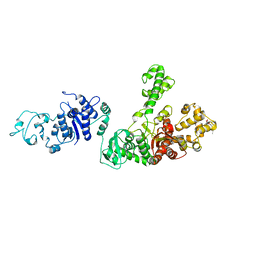 | | STRUCTURE OF TAQ DNA POLYMERASE | | Descriptor: | 2-O-octyl-beta-D-glucopyranose, TAQ DNA POLYMERASE, ZINC ION | | Authors: | Kim, Y, Eom, S.H, Wang, J, Lee, D.-S, Suh, S.W, Steitz, T.A. | | Deposit date: | 1996-06-04 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Thermus aquaticus DNA polymerase.
Nature, 376, 1995
|
|
3SFP
 
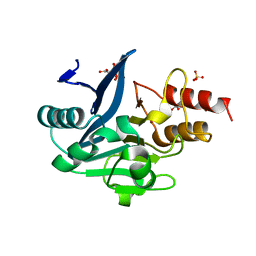 | | Crystal Structure of the Mono-Zinc-boundform of New Delhi Metallo-beta-Lactamase-1 from Klebsiella pneumoniae | | Descriptor: | Beta-lactamase NDM-1, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2011-06-13 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure of Apo- and Monometalated Forms of NDM-1 A Highly Potent Carbapenem-Hydrolyzing Metallo-beta-Lactamase
Plos One, 6, 2011
|
|
3SQM
 
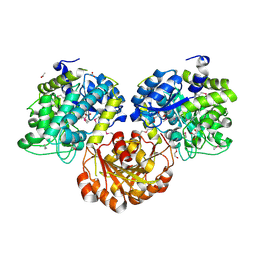 | | Crystal Structure of Glycoside Hydrolase from Synechococcus Complexed with N-acetyl-D-glucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-07-05 | | Release date: | 2011-07-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal Structure of Glycoside Hydrolase from Synechococcus Complexed with N-acetyl-D-glucosamine
To be Published
|
|
3SRX
 
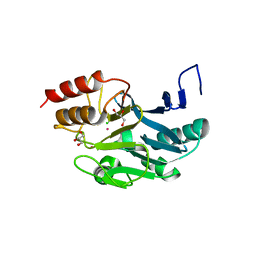 | | New Delhi Metallo-beta-Lactamase-1 Complexed with Cd | | Descriptor: | Beta-lactamase NDM-1, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2011-07-07 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New Delhi Metallo-beta-Lactamase-1 Complexed with Cd
To be Published
|
|
3SQL
 
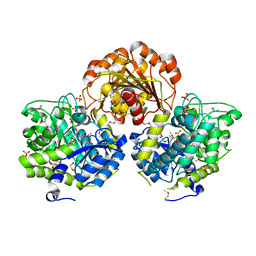 | | Crystal Structure of Glycoside Hydrolase from Synechococcus | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-07-05 | | Release date: | 2011-07-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Glycoside Hydrolase from Synechococcus
To be Published
|
|
3SBL
 
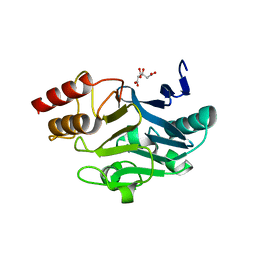 | | Crystal Structure of New Delhi Metal-beta-lactamase-1 from Klebsiella pneumoniae | | Descriptor: | Beta-lactamase NDM-1, CITRIC ACID | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2011-06-05 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of Apo- and Monometalated Forms of NDM-1 A Highly Potent Carbapenem-Hydrolyzing Metallo-beta-Lactamase
Plos One, 6, 2011
|
|
3TVA
 
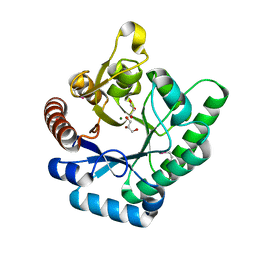 | | Crystal Structure of Xylose isomerase domain protein from Planctomyces limnophilus | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kim, Y, Wu, R, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-19 | | Release date: | 2011-10-05 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Crystal Structure of Xylose isomerase domain protein from Planctomyces limnophilus
To be Published
|
|
3TY6
 
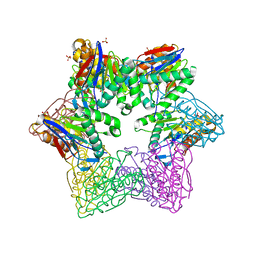 | | ATP-dependent Protease HslV from Bacillus anthracis str. Ames | | Descriptor: | ATP-dependent protease subunit HslV, SULFATE ION | | Authors: | Kim, Y, Mulligan, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-23 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | ATP-dependent Protease HslV from Bacillus anthracis str. Ames
To be Published
|
|
