1OHT
 
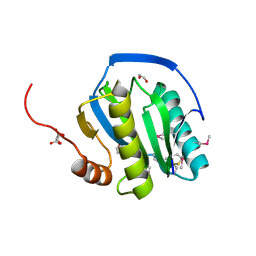 | | Peptidoglycan recognition protein LB | | 分子名称: | 1,2-ETHANEDIOL, CG14704 PROTEIN, L(+)-TARTARIC ACID, ... | | 著者 | Kim, M.-S, Byun, M, Oh, B.-H. | | 登録日 | 2003-05-31 | | 公開日 | 2003-07-18 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of Peptidoglycan Recognition Protein Lb from Drosophila Melanogaster
Nat.Immunol., 4, 2003
|
|
2B7Q
 
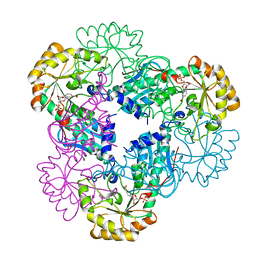 | | Crystal structure of quinolinic acid phosphoribosyltransferase from Helicobacter pylori with nicotinate mononucleotide | | 分子名称: | NICOTINATE MONONUCLEOTIDE, Probable nicotinate-nucleotide pyrophosphorylase | | 著者 | Kim, M.K, Im, Y.J, Lee, J.H, Eom, S.H. | | 登録日 | 2005-10-05 | | 公開日 | 2006-02-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | Crystal structure of quinolinic acid phosphoribosyltransferase from Helicobacter pylori
Proteins, 63, 2006
|
|
2B7P
 
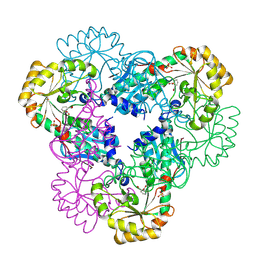 | | Crystal structure of quinolinic acid phosphoribosyltransferase from Helicobacter pylori with phthalic acid | | 分子名称: | PHTHALIC ACID, Probable nicotinate-nucleotide pyrophosphorylase, SULFATE ION | | 著者 | Kim, M.K, Im, Y.J, Lee, J.H, Eom, S.H. | | 登録日 | 2005-10-05 | | 公開日 | 2006-02-14 | | 最終更新日 | 2018-09-19 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Crystal structure of quinolinic acid phosphoribosyltransferase from Helicobacter pylori
Proteins, 63, 2006
|
|
2B7N
 
 | | Crystal structure of quinolinic acid phosphoribosyltransferase from Helicobacter pylori | | 分子名称: | Probable nicotinate-nucleotide pyrophosphorylase, QUINOLINIC ACID, SULFATE ION | | 著者 | Kim, M.K, Im, Y.J, Lee, J.H, Eom, S.H. | | 登録日 | 2005-10-04 | | 公開日 | 2006-02-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of quinolinic acid phosphoribosyltransferase from Helicobacter pylori
Proteins, 63, 2006
|
|
1MJD
 
 | | Structure of N-terminal domain of human doublecortin | | 分子名称: | DOUBLECORTIN | | 著者 | Kim, M.H, Cierpicki, T, Derewenda, U, Krowarsch, D, Feng, Y, Devedjiev, Y, Dauter, Z, Walsh, C.A, Otlewski, J, Bushweller, J.H, Derewenda, Z.S. | | 登録日 | 2002-08-27 | | 公開日 | 2003-04-29 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The DCX-domain Tandems of Doublecortin and Doublecortin-like Kinase
Nat.Struct.Biol., 10, 2003
|
|
1OGF
 
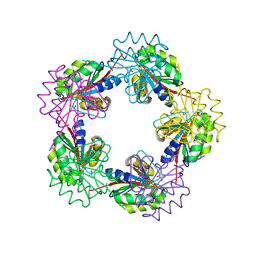 | |
1OGE
 
 | |
1OGC
 
 | |
1OGD
 
 | |
6K40
 
 | | Crystal structure of alkyl hydroperoxide reductase from D. radiodurans R1 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alkyl hydroperoxide reductase AhpD, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Kim, M.-K, Zhang, J, Zhao, L. | | 登録日 | 2019-05-22 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Crystal structure of the AhpD-like protein DR1765 from Deinococcus radiodurans R1.
Biochem.Biophys.Res.Commun., 529, 2020
|
|
6IZ2
 
 | |
4OOZ
 
 | | Crystal structure of beta-1,4-D-mannanase from Cryptopygus antarcticus in complex with mannopentaose | | 分子名称: | Beta-1,4-mannanase, beta-D-mannopyranose, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, ... | | 著者 | Kim, M.-K, An, Y.J, Jeong, C.-S, Cha, S.-S. | | 登録日 | 2014-02-04 | | 公開日 | 2014-08-06 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure-based investigation into the functional roles of the extended loop and substrate-recognition sites in an endo-beta-1,4-d-mannanase from the Antarctic springtail, Cryptopygus antarcticus.
Proteins, 82, 2014
|
|
4FEC
 
 | | Crystal Structure of Htt36Q3H | | 分子名称: | Maltose-binding periplasmic protein,Huntingtin, ZINC ION | | 著者 | Kim, M. | | 登録日 | 2012-05-30 | | 公開日 | 2013-03-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues.
Prion, 7, 2013
|
|
4FEB
 
 | | Crystal Structure of Htt36Q3H-EX1-X1-C2(Beta) | | 分子名称: | Maltose-binding periplasmic protein,Huntingtin, SODIUM ION, ZINC ION | | 著者 | Kim, M. | | 登録日 | 2012-05-29 | | 公開日 | 2013-03-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues.
Prion, 7, 2013
|
|
4FE8
 
 | | Crystal Structure of Htt36Q3H-EX1-X1-C1(Alpha) | | 分子名称: | Maltose-binding periplasmic protein,Huntingtin, ZINC ION | | 著者 | Kim, M. | | 登録日 | 2012-05-29 | | 公開日 | 2013-03-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues.
Prion, 7, 2013
|
|
4FED
 
 | | Crystal Structure of Htt36Q3H | | 分子名称: | CALCIUM ION, Maltose-binding periplasmic protein,Huntingtin, ZINC ION | | 著者 | Kim, M. | | 登録日 | 2012-05-30 | | 公開日 | 2013-03-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.807 Å) | | 主引用文献 | Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues.
Prion, 7, 2013
|
|
7TM3
 
 | | Structure of the rabbit 80S ribosome stalled on a 2-TMD Rhodopsin intermediate in complex with the multipass translocon | | 分子名称: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | 著者 | Kim, M.K, Lewis, A.J.O, Keenan, R.J, Hegde, R.S. | | 登録日 | 2022-01-19 | | 公開日 | 2022-10-19 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | Mechanism of an intramembrane chaperone for multipass membrane proteins.
Nature, 611, 2022
|
|
7TUT
 
 | | Structure of the rabbit 80S ribosome stalled on a 4-TMD Rhodopsin intermediate in complex with the multipass translocon | | 分子名称: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | 著者 | Kim, M.K, Lewis, A.J.O, Keenan, R.J, Hegde, R.S. | | 登録日 | 2022-02-03 | | 公開日 | 2022-10-19 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.88 Å) | | 主引用文献 | Mechanism of an intramembrane chaperone for multipass membrane proteins.
Nature, 611, 2022
|
|
4EJ0
 
 | |
4OOU
 
 | | Crystal structure of beta-1,4-D-mannanase from Cryptopygus antarcticus | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-1,4-mannanase | | 著者 | Kim, M.-K, An, Y.J, Jeong, C.-S, Cha, S.-S. | | 登録日 | 2014-02-04 | | 公開日 | 2014-08-06 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Structure-based investigation into the functional roles of the extended loop and substrate-recognition sites in an endo-beta-1,4-d-mannanase from the Antarctic springtail, Cryptopygus antarcticus.
Proteins, 82, 2014
|
|
1IVW
 
 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Late intermediate in topaquinone biogenesis | | 分子名称: | COPPER (II) ION, amine oxidase | | 著者 | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | 登録日 | 2002-03-29 | | 公開日 | 2002-08-07 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
4J4K
 
 | | Crystal structure of glucose isomerase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Xylose isomerase, ... | | 著者 | Kim, M.K, An, Y.J, Lee, S, Jeong, C.S, Cha, S.S. | | 登録日 | 2013-02-07 | | 公開日 | 2014-04-30 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of glucose isomerase
To be Published
|
|
1IVV
 
 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Early intermediate in topaquinone biogenesis | | 分子名称: | COPPER (II) ION, amine oxidase | | 著者 | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | 登録日 | 2002-03-29 | | 公開日 | 2002-08-07 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
1IVU
 
 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Initial intermediate in topaquinone biogenesis | | 分子名称: | COPPER (II) ION, amine oxidase | | 著者 | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | 登録日 | 2002-03-29 | | 公開日 | 2002-08-07 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
5HYY
 
 | |
