4L4H
 
 | |
4I2S
 
 | |
4I8M
 
 | |
4I7I
 
 | |
4I96
 
 | |
4I1E
 
 | |
4I3N
 
 | |
4I37
 
 | |
5IO3
 
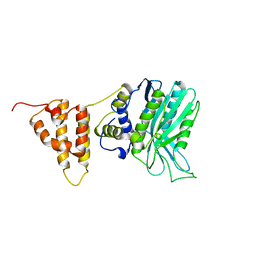 | | Crystal structure of the legionella pneumophila effector protein RavZ - I422 | | Descriptor: | Uncharacterized protein RavZ | | Authors: | Kwon, D.H, Kim, L, Kim, B.-W, Hong, S.B, Song, H.K. | | Deposit date: | 2016-03-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane
Autophagy, 13, 2017
|
|
7P81
 
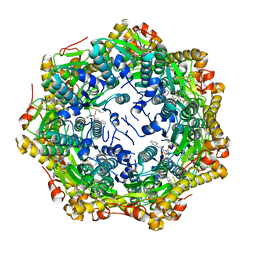 | | Crystal structure of ClpP from Bacillus subtilis in complex with ADEP2 (compact state) | | Descriptor: | ADEP2, ATP-dependent Clp protease proteolytic subunit | | Authors: | Lee, B.-G, Kim, L, Kim, M.K, Kwon, D.H, Song, H.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
7P80
 
 | | Crystal structure of ClpP from Bacillus subtilis in complex with ADEP2 (compressed state) | | Descriptor: | ADEP2, ATP-dependent Clp protease proteolytic subunit | | Authors: | Lee, B.-G, Kim, L, Kim, M.K, Kwon, D.H, Song, H.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
2PS3
 
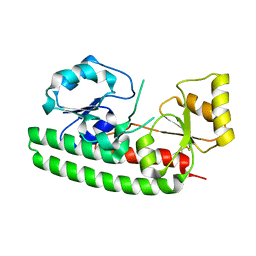 | | Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli | | Descriptor: | High-affinity zinc uptake system protein znuA | | Authors: | Yatsunyk, L.A, Kim, L.R, Vorontsov, I.I, Rosenzweig, A.C. | | Deposit date: | 2007-05-04 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli.
J.Biol.Inorg.Chem., 13, 2008
|
|
7D34
 
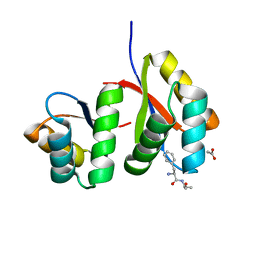 | | AtClpS1-peptide complex | | Descriptor: | ACETIC ACID, ALANINE, ATP-dependent Clp protease adapter protein CLPS1, ... | | Authors: | Heo, J, Kim, L, Kwon, D.H, Song, H.K. | | Deposit date: | 2020-09-18 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structural basis for the N-degron specificity of ClpS1 from Arabidopsis thaliana.
Protein Sci., 30, 2021
|
|
5IZV
 
 | | Crystal structure of the legionella pneumophila effector protein RavZ - F222 | | Descriptor: | Uncharacterized protein RavZ | | Authors: | Kwon, D.H, Kim, L, Kim, B.-W, Hong, S.B, Song, H.K. | | Deposit date: | 2016-03-26 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.814 Å) | | Cite: | The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane
Autophagy, 13, 2017
|
|
5HZY
 
 | | Crystal structure of the legionella pneumophila effector protein RavZ - P6322 | | Descriptor: | Uncharacterized protein RavZ | | Authors: | Kwon, D.H, Kim, L, Kim, B.-W, Hong, S.B, Song, H.K. | | Deposit date: | 2016-02-03 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane
Autophagy, 13, 2017
|
|
2PRS
 
 | | Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli | | Descriptor: | High-affinity zinc uptake system protein znuA, ISOPROPYL ALCOHOL, ZINC ION | | Authors: | Yatsunyk, L.A, Kim, L.R, Vorontsov, I.I, Rosenzweig, A.C. | | Deposit date: | 2007-05-04 | | Release date: | 2007-06-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli.
J.Biol.Inorg.Chem., 13, 2008
|
|
2PS0
 
 | | Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli | | Descriptor: | High-affinity zinc uptake system protein znuA, ZINC ION | | Authors: | Yatsunyk, L.A, Kim, L.R, Vorontsov, I.I, Rosenzweig, A.C. | | Deposit date: | 2007-05-04 | | Release date: | 2007-06-05 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli.
J.Biol.Inorg.Chem., 13, 2008
|
|
2PS9
 
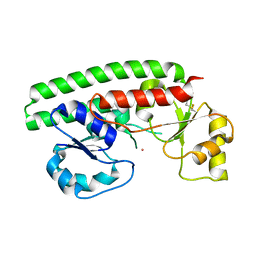 | | Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli | | Descriptor: | COBALT (II) ION, High-affinity zinc uptake system protein znuA | | Authors: | Yatsunyk, L.A, Kim, L.R, Vorontsov, I.I, Rosenzweig, A.C. | | Deposit date: | 2007-05-04 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli.
J.Biol.Inorg.Chem., 13, 2008
|
|
5XAC
 
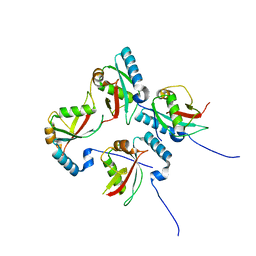 | | CLIR - LC3B | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-03-12 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | A novel conformation of the LC3-interacting region motif revealed by the structure of a complex between LC3B and RavZ
Biochem. Biophys. Res. Commun., 490, 2017
|
|
5XAE
 
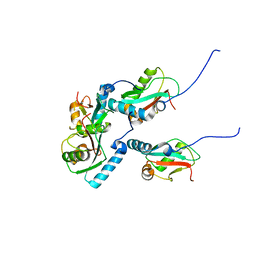 | | mutNLIR_LC3B | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-03-12 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | A novel conformation of the LC3-interacting region motif revealed by the structure of a complex between LC3B and RavZ
Biochem. Biophys. Res. Commun., 490, 2017
|
|
5XAD
 
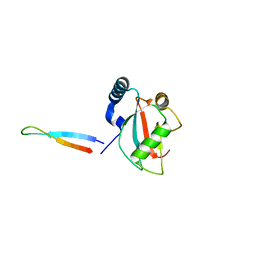 | | NLIR - LC3B fusion protein | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, Uncharacterised protein | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-03-12 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A novel conformation of the LC3-interacting region motif revealed by the structure of a complex between LC3B and RavZ
Biochem. Biophys. Res. Commun., 490, 2017
|
|
5YPC
 
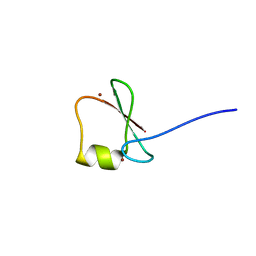 | | p62/SQSTM1 ZZ domain with Phe-peptide | | Descriptor: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-11-01 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
5YPB
 
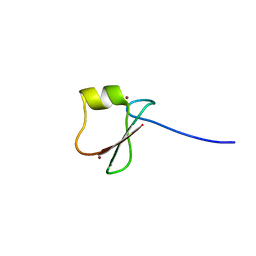 | | p62/SQSTM1 ZZ domain with His-peptide | | Descriptor: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-11-01 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
5YPF
 
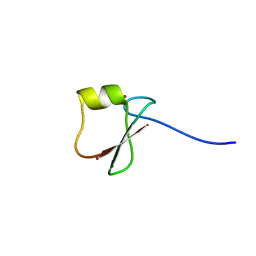 | | p62/SQSTM1 ZZ domain with Trp-peptide | | Descriptor: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-11-01 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
5YPE
 
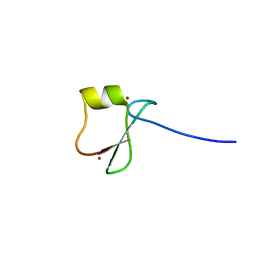 | | p62/SQSTM1 ZZ domain with Tyr-peptide | | Descriptor: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-11-01 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
