3F22
 
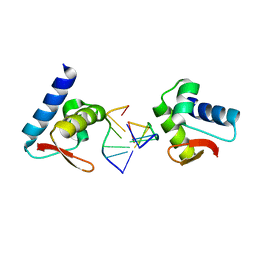 | | Crystal structure of Zalpha in complex with d(CGTACG) | | Descriptor: | DNA (5'-D(*DTP*DCP*DGP*DTP*DAP*DCP*DG)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Ha, S.C, Choi, J, Kim, K.K. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structures of non-CG-repeat Z-DNAs co-crystallized with the Z-DNA-binding domain, hZ{alpha}ADAR1
Nucleic Acids Res., 37, 2009
|
|
2EOT
 
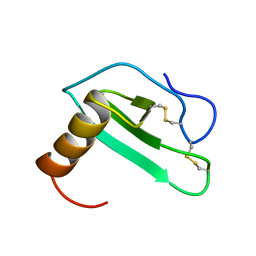 | | SOLUTION STRUCTURE OF EOTAXIN, AN ENSEMBLE OF 32 NMR SOLUTION STRUCTURES | | Descriptor: | EOTAXIN | | Authors: | Crump, M.P, Rajarathnam, K, Kim, K.-S, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 1998-06-29 | | Release date: | 1998-11-11 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of eotaxin, a chemokine that selectively recruits eosinophils in allergic inflammation.
J.Biol.Chem., 273, 1998
|
|
3S8R
 
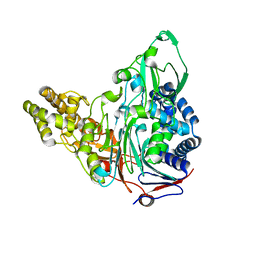 | | Crystal Structures of Glutaryl 7-Aminocephalosporanic Acid Acylase: Insight into Autoproteolytic Activation | | Descriptor: | GLYCEROL, Glutaryl-7-aminocephalosporanic-acid acylase | | Authors: | Kim, J.K, Yang, I.S, Park, S.S, Kim, K.H. | | Deposit date: | 2011-05-30 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of glutaryl 7-aminocephalosporanic acid acylase: insight into autoproteolytic activation.
Biochemistry, 42, 2003
|
|
1OR0
 
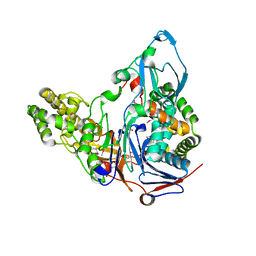 | | Crystal Structures of Glutaryl 7-Aminocephalosporanic Acid Acylase: Insight into Autoproteolytic Activation | | Descriptor: | 1,2-ETHANEDIOL, Glutaryl 7-Aminocephalosporanic Acid Acylase, glutaryl acylase | | Authors: | Kim, J.K, Yang, I.S, Rhee, S, Dauter, Z, Lee, Y.S, Park, S.S, Kim, K.H. | | Deposit date: | 2003-03-11 | | Release date: | 2004-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Glutaryl 7-Aminocephalosporanic Acid Acylase: Insight into Autoproteolytic Activation
Biochemistry, 42, 2003
|
|
1NM3
 
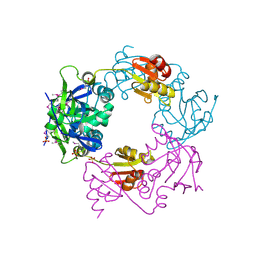 | | Crystal structure of Heamophilus influenza hybrid-Prx5 | | Descriptor: | Protein HI0572, SULFATE ION | | Authors: | Kim, S.J, Woo, J.R, Hwang, Y.S, Jeong, D.G, Shin, D.H, Kim, K.H, Ryu, S.E. | | Deposit date: | 2003-01-08 | | Release date: | 2003-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Tetrameric Structure of Haemophilus influenza Hybrid Prx5 Reveals Interactions between Electron Donor and Acceptor Proteins.
J.Biol.Chem., 278, 2003
|
|
7D7O
 
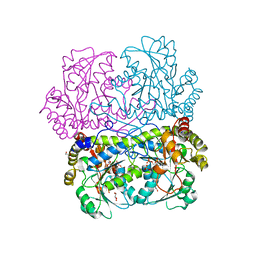 | | Crystal structure of cystathionine gamma-lyase from Bacillus cereus ATCC 14579 | | Descriptor: | Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Sagong, H.-Y, Kim, B, Kim, K.-J. | | Deposit date: | 2020-10-05 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural and Functional Characterization of Cystathionine gamma-lyase from Bacillus cereus ATCC 14579.
J.Agric.Food Chem., 68, 2020
|
|
4PRL
 
 | |
4P85
 
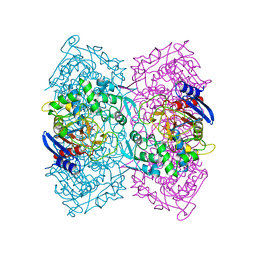 | | Crystal structure of Est-Y29, a novel penicillin-binding protein/beta-lactamase homolog from a metagenomic library | | Descriptor: | DIETHYL PHOSPHONATE, Est-Y29 | | Authors: | Ngo, T.D, Ryu, B.H, Ju, H.S, Jang, E.J, Kim, K.K, Kim, D.H. | | Deposit date: | 2014-03-30 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic analysis and biochemical applications of a novel penicillin-binding protein/ beta-lactamase homologue from a metagenomic library.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PRK
 
 | |
1NAG
 
 | | CREVICE-FORMING MUTANTS IN THE RIGID CORE OF BOVINE PANCREATIC TRYPSIN INHIBITOR: CRYSTAL STRUCTURES OF F22A, Y23A, N43G, AND F45A | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR, PHOSPHATE ION | | Authors: | Danishefsky, A.T, Wlodawer, A, Kim, K.-S, Tao, F, Woodward, C. | | Deposit date: | 1992-08-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crevice-forming mutants in the rigid core of bovine pancreatic trypsin inhibitor: crystal structures of F22A, Y23A, N43G, and F45A.
Protein Sci., 2, 1993
|
|
4O9A
 
 | | Crystal structure of Beta-ketothiolase (PhaA) from Ralstonia eutropha H16 | | Descriptor: | Acetyl-CoA acetyltransferase | | Authors: | Kim, E.J, Kim, J, Kim, S, Kim, K.J. | | Deposit date: | 2014-01-02 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structure and biochemical characterization of PhaA from Ralstonia eutropha, a polyhydroxyalkanoate-producing bacterium.
Biochem.Biophys.Res.Commun., 452, 2014
|
|
4P6B
 
 | | Crystal structure of Est-Y29,a novel penicillin-binding protein/beta-lactamase homolog from a metagenomic library | | Descriptor: | Est-Y29 | | Authors: | Ngo, T.D, Ryu, B.H, Ju, H.S, Jang, E.J, Kim, K.K, Kim, D.H. | | Deposit date: | 2014-03-24 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic analysis and biochemical applications of a novel penicillin-binding protein/ beta-lactamase homologue from a metagenomic library.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3R4Y
 
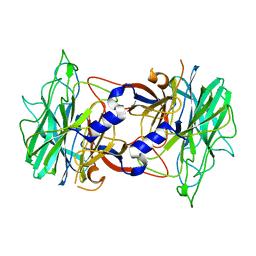 | | Crystal structure of alpha-neoagarobiose hydrolase (ALPHA-NABH) from Saccharophagus degradans 2-40 | | Descriptor: | Glycosyl hydrolase family 32, N terminal | | Authors: | Lee, S, Lee, J.Y, Ha, S.C, Shin, D.H, Kim, K.H, Bang, W.G, Kim, S.H, Choi, I.G. | | Deposit date: | 2011-03-18 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a key enzyme in the agarolytic pathway, alpha-neoagarobiose hydrolase from Saccharophagus degradans 2-40
Biochem.Biophys.Res.Commun., 412, 2011
|
|
7CVW
 
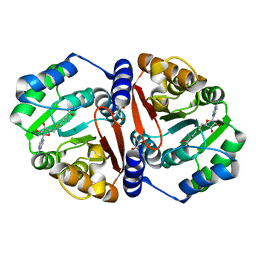 | |
7CVV
 
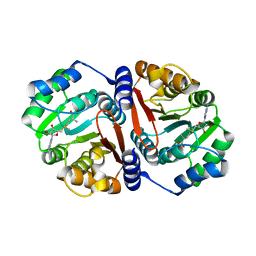 | |
7CVU
 
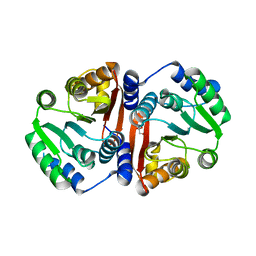 | |
4O99
 
 | | Crystal structure of Beta-ketothiolase (PhaA) from Ralstonia eutropha H16 | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL | | Authors: | Kim, E.J, Kim, J, Kim, S, Kim, K.J. | | Deposit date: | 2014-01-02 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure and biochemical characterization of PhaA from Ralstonia eutropha, a polyhydroxyalkanoate-producing bacterium.
Biochem.Biophys.Res.Commun., 452, 2014
|
|
4P87
 
 | | Crystal structure of Est-Y29, a novel penicillin-binding protein/beta-lactamase homolog from a metagenomic library | | Descriptor: | 4-NITROPHENYL PHOSPHATE, Est-Y29 | | Authors: | Ngo, T.D, Ryu, B.H, Ju, H.S, Jang, E.J, Kim, K.K, Kim, D.H. | | Deposit date: | 2014-03-30 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Crystallographic analysis and biochemical applications of a novel penicillin-binding protein/ beta-lactamase homologue from a metagenomic library.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3R4Z
 
 | | Crystal structure of alpha-neoagarobiose hydrolase (ALPHA-NABH) in complex with alpha-d-galactopyranose from Saccharophagus degradans 2-40 | | Descriptor: | Glycosyl hydrolase family 32, N terminal, alpha-D-galactopyranose | | Authors: | Lee, S, Lee, J.Y, Ha, S.C, Shin, D.H, Kim, K.H, Bang, W.G, Kim, S.H, Choi, I.G. | | Deposit date: | 2011-03-18 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of a key enzyme in the agarolytic pathway, alpha-neoagarobiose hydrolase from Saccharophagus degradans 2-40
Biochem.Biophys.Res.Commun., 412, 2011
|
|
1MZL
 
 | | MAIZE NONSPECIFIC LIPID TRANSFER PROTEIN | | Descriptor: | MAIZE NONSPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Shin, D.H, Lee, J.Y, Hwang, K.Y, Kim, K.K, Suh, S.W. | | Deposit date: | 1995-01-26 | | Release date: | 1996-08-01 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structure of the non-specific lipid-transfer protein from maize seedlings.
Structure, 3, 1995
|
|
7ETX
 
 | |
7ETY
 
 | |
7VGM
 
 | |
1EFE
 
 | | AN ACTIVE MINI-PROINSULIN, M2PI | | Descriptor: | MINI-PROINSULIN | | Authors: | Cho, Y, Chang, S.G, Choi, K.D, Shin, H, Ahn, B, Kim, K.S. | | Deposit date: | 2000-02-08 | | Release date: | 2000-03-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of an Active Mini-Proinsulin, M2PI: Inter-chain Flexibility is Crucial for Insulin Activity
J.Biochem.Mol.Biol., 33, 2000
|
|
4YGR
 
 | | Crystal structure of HAD phosphatase from Thermococcus onnurineus | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hydrolase, MAGNESIUM ION | | Authors: | Ngo, T.D, Le, B.V, Subramani, V.K, Nguyen, C.M.T, Lee, H.S, Cho, Y, Kim, K.K, Hwang, H.Y. | | Deposit date: | 2015-02-26 | | Release date: | 2015-04-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Structural basis for the substrate selectivity of a HAD phosphatase from Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 461, 2015
|
|
