4QNU
 
 | | Crystal structure of CmoB bound with Cx-SAM in P21212 | | Descriptor: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, PHOSPHATE ION, tRNA (mo5U34)-methyltransferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-06-18 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Determinants of the CmoB carboxymethyl transferase utilized for selective tRNA wobble modification.
Nucleic Acids Res., 43, 2015
|
|
7EQJ
 
 | | crystal structure of E. coli Valine tRNA | | Descriptor: | MAGNESIUM ION, RNA (76-MER), SODIUM ION | | Authors: | Kim, J, Jeong, H. | | Deposit date: | 2021-05-03 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Unique anticodon loop conformation with the flipped-out wobble nucleotide in the crystal structure of unbound tRNA Val .
Rna, 27, 2021
|
|
4GRA
 
 | | Crystal structure of SULT1A1 bound with PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase 1A1 | | Authors: | Kim, J, Cook, I, Wang, T, Falany, C.N, Leyh, T.S, Almo, S.C. | | Deposit date: | 2012-08-24 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The gate that governs sulfotransferase selectivity.
Biochemistry, 52, 2013
|
|
6L5H
 
 | |
4IFB
 
 | | Crystal structure of SULT 2A1 LLGG mutant with PAPS | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-PHOSPHATE SULFATE, Bile salt sulfotransferase, PHOSPHATE ION, ... | | Authors: | Kim, J, Toro, R, Bhosle, R, Cook, I, Wang, T, Falany, C.N, Leyh, T.S, Almo, S.C. | | Deposit date: | 2012-12-14 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sulfotransferase Selectivity at the Molecular level
To be Published
|
|
6L5J
 
 | |
4KDZ
 
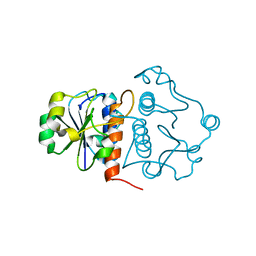 | |
5ZIQ
 
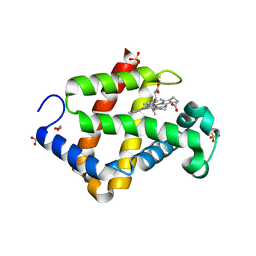 | | Crystal structure of hexacoordinated heme protein from anhydrobiotic tardigrade at pH 4 | | Descriptor: | 1,2-ETHANEDIOL, Globin protein, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kim, J, Fukuda, Y, Inoue, T. | | Deposit date: | 2018-03-16 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Kumaglobin: a hexacoordinated heme protein from an anhydrobiotic tardigrade, Ramazzottius varieornatus.
FEBS J., 286, 2019
|
|
5ZM9
 
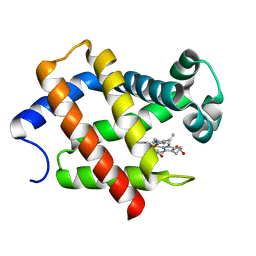 | | Crystal structure of hexacoordinated heme protein from anhydrobiotic tardigrade at pH 7 | | Descriptor: | CHLORIDE ION, Globin Protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kim, J, Fukuda, Y, Inoue, T. | | Deposit date: | 2018-04-02 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Kumaglobin: a hexacoordinated heme protein from an anhydrobiotic tardigrade, Ramazzottius varieornatus.
FEBS J., 286, 2019
|
|
3OCQ
 
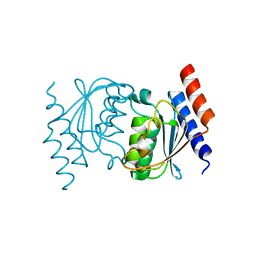 | |
3ODG
 
 | | crystal structure of xanthosine phosphorylase bound with xanthine from Yersinia pseudotuberculosis | | Descriptor: | CHLORIDE ION, XANTHINE, Xanthosine phosphorylase | | Authors: | Kim, J, Ramagopal, U.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-08-11 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | crystal structure of xanthosine phosphorylase bound with xanthine from Yersinia pseudotuberculosis
To be Published
|
|
2OB3
 
 | | Structure of Phosphotriesterase mutant H257Y/L303T | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Parathion hydrolase, ZINC ION | | Authors: | Kim, J, Ramagopal, U.A, Tsai, P, Raushel, F.M, Almo, S.C. | | Deposit date: | 2006-12-18 | | Release date: | 2007-01-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structure of Phosphotriesterase mutant H257Y/L303T
To be Published
|
|
2O4M
 
 | | Structure of Phosphotriesterase mutant I106G/F132G/H257Y | | Descriptor: | ACETIC ACID, CACODYLATE ION, GLYCEROL, ... | | Authors: | Kim, J, Ramagopal, U.A, Tsai, P, Raushel, F.M, Almo, S.C. | | Deposit date: | 2006-12-04 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of Phosphotriesterase mutant I106G/F132G/H257Y
To be Published
|
|
2O4Q
 
 | | Structure of Phosphotriesterase mutant G60A | | Descriptor: | CACODYLATE ION, Parathion hydrolase, ZINC ION | | Authors: | Kim, J, Ramagopal, U.A, Tsai, P.C, Raushel, F.M, Almo, S.C. | | Deposit date: | 2006-12-04 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of diethyl phosphate bound to the binuclear metal center of phosphotriesterase.
Biochemistry, 47, 2008
|
|
3OF3
 
 | | Crystal structure of PNP with an inhibitor DADME_immH from Vibrio cholerae | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase deoD-type 1 | | Authors: | Kim, J, Ramagopal, U.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-08-13 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of PNP with an inhibitor DADME_immH from Vibrio cholerae
To be Published
|
|
3OCC
 
 | | Crystal structure of PNP with DADMEimmH from Yersinia pseudotuberculosis | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase deoD-type | | Authors: | Kim, J, Ramagopal, U.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-08-09 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | crystal structure of PNP with DADMEimmH from Yersinia pseudotuberculosis
To be Published
|
|
3OHP
 
 | |
3PVC
 
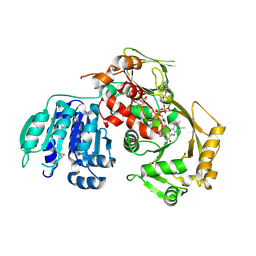 | |
3PS9
 
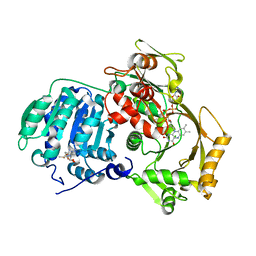 | | Crystal structure of MnmC from E. coli | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, S-ADENOSYLMETHIONINE, ... | | Authors: | Kim, J, Almo, S.C. | | Deposit date: | 2010-12-01 | | Release date: | 2010-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural basis for hypermodification of the wobble uridine in tRNA by bifunctional enzyme MnmC.
Bmc Struct.Biol., 13, 2013
|
|
3RCM
 
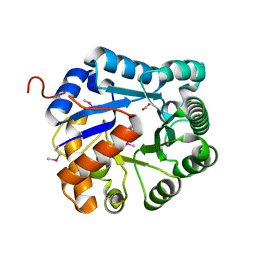 | | crystal structure of EFI target 500140:TatD family hydrolase from Pseudomonas putida | | Descriptor: | ACETATE ION, CITRIC ACID, TatD family hydrolase, ... | | Authors: | Kim, J, Toro, R, Hillerich, B, Seidel, R.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-03-31 | | Release date: | 2011-04-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | crystal structure of EFI target 500140:TatD family hydrolase from Pseudomonas putida
TO BE PUBLISHED
|
|
3SGL
 
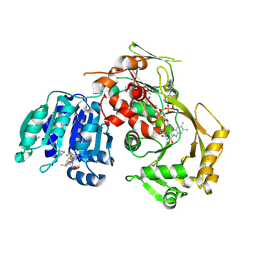 | |
1K5U
 
 | |
1K5V
 
 | |
5X06
 
 | | DNA replication regulation protein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA polymerase III subunit beta, DnaA regulatory inactivator Hda, ... | | Authors: | Kim, J, Cho, Y. | | Deposit date: | 2017-01-20 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.237 Å) | | Cite: | Replication regulation protein
To Be Published
|
|
5ZW3
 
 | | Crystal Structure of TrmR from B. subtilis | | Descriptor: | MAGNESIUM ION, Putative O-methyltransferase YrrM, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Kim, J, Ryu, H, Almo, S.C. | | Deposit date: | 2018-05-14 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Identification of a novel tRNA wobble uridine modifying activity in the biosynthesis of 5-methoxyuridine.
Nucleic Acids Res., 46, 2018
|
|
