8EBV
 
 | |
8EBY
 
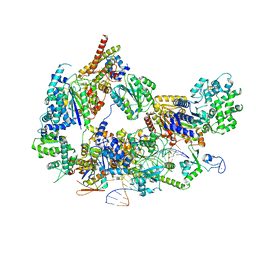 | | XPC release from Core7-XPA-DNA (AP) | | Descriptor: | DNA, DNA repair protein complementing XP-A cells, DNA repair protein complementing XP-C cells, ... | | Authors: | Kim, J, Yang, W. | | Deposit date: | 2022-08-31 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Lesion recognition by XPC, TFIIH and XPA in DNA excision repair.
Nature, 617, 2023
|
|
8EBS
 
 | |
8EBT
 
 | |
8EBW
 
 | |
8EBX
 
 | |
6ICQ
 
 | | Pseudomonas putida CBB5 NdmA QL mutant with theobromine | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, Methylxanthine N1-demethylase NdmA, ... | | Authors: | Kim, J.H, Kim, B.H, Kang, S.Y, Song, H.K. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Mechanistic Insights into Caffeine Degradation by the Bacterial N-Demethylase Complex.
J.Mol.Biol., 431, 2019
|
|
3MDD
 
 | |
6ICN
 
 | | Pseudomonas putida CBB5 NdmA with caffeine | | Descriptor: | CAFFEINE, COBALT (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Kim, J.H, Kim, B.H, Kang, S.Y, Song, H.K. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Mechanistic Insights into Caffeine Degradation by the Bacterial N-Demethylase Complex.
J.Mol.Biol., 431, 2019
|
|
1MIO
 
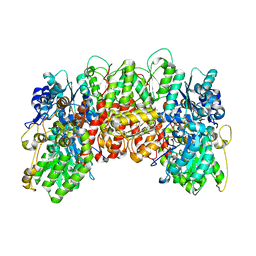 | | X-RAY CRYSTAL STRUCTURE OF THE NITROGENASE MOLYBDENUM-IRON PROTEIN FROM CLOSTRIDIUM PASTEURIANUM AT 3.0 ANGSTROMS RESOLUTION | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE-MO-S CLUSTER, ... | | Authors: | Kim, J, Woo, D, Rees, D.C. | | Deposit date: | 1993-03-24 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray crystal structure of the nitrogenase molybdenum-iron protein from Clostridium pasteurianum at 3.0-A resolution.
Biochemistry, 32, 1993
|
|
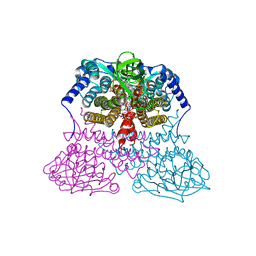
3MDE
 
 | |
6ICO
 
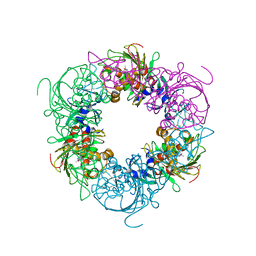 | | Pseudomonas putida CBB5 NdmA with theophylline | | Descriptor: | COBALT (II) ION, FE2/S2 (INORGANIC) CLUSTER, Methylxanthine N1-demethylase NdmA, ... | | Authors: | Kim, J.H, Kim, B.H, Kang, S.Y, Song, H.K. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Mechanistic Insights into Caffeine Degradation by the Bacterial N-Demethylase Complex.
J.Mol.Biol., 431, 2019
|
|
6ICM
 
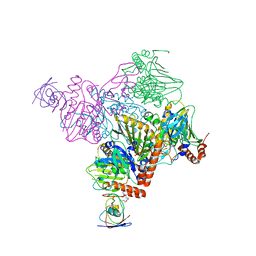 | | Pseudomonas putida CBB5 NdmA with ferredoxin domain of NdmD | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, Methylxanthine N1-demethylase NdmA, ... | | Authors: | Kim, J.H, Kim, B.H, Kang, S.Y, Song, H.K. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.961 Å) | | Cite: | Structural and Mechanistic Insights into Caffeine Degradation by the Bacterial N-Demethylase Complex.
J.Mol.Biol., 431, 2019
|
|
6ICK
 
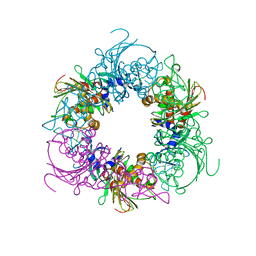 | | Pseudomonas putida CBB5 NdmA | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, Methylxanthine N1-demethylase NdmA | | Authors: | Kim, J.H, Kim, B.H, Kang, S.Y, Song, H.K. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Structural and Mechanistic Insights into Caffeine Degradation by the Bacterial N-Demethylase Complex.
J.Mol.Biol., 431, 2019
|
|
6JDK
 
 | |
2PNV
 
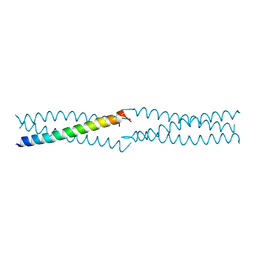 | | Crystal Structure of the leucine zipper domain of small-conductance Ca2+-activated K+ (SKCa) channel from Rattus norvegicus | | Descriptor: | Small conductance calcium-activated potassium channel protein 2 | | Authors: | Kim, J.Y, Kim, M.K, Kang, G.B, Park, C.S, Eom, S.H. | | Deposit date: | 2007-04-25 | | Release date: | 2008-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the leucine zipper domain of small-conductance Ca2+-activated K+ (SK(Ca)) channel from Rattus norvegicus.
Proteins, 70, 2008
|
|
1OR0
 
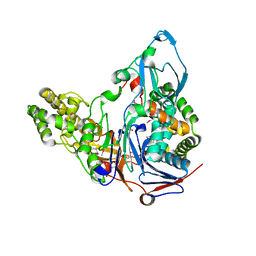 | | Crystal Structures of Glutaryl 7-Aminocephalosporanic Acid Acylase: Insight into Autoproteolytic Activation | | Descriptor: | 1,2-ETHANEDIOL, Glutaryl 7-Aminocephalosporanic Acid Acylase, glutaryl acylase | | Authors: | Kim, J.K, Yang, I.S, Rhee, S, Dauter, Z, Lee, Y.S, Park, S.S, Kim, K.H. | | Deposit date: | 2003-03-11 | | Release date: | 2004-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Glutaryl 7-Aminocephalosporanic Acid Acylase: Insight into Autoproteolytic Activation
Biochemistry, 42, 2003
|
|
6ICL
 
 | | Pseudomonas putida CBB5 NdmB | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, Methylxanthine N3-demethylase NdmB | | Authors: | Kim, J.H, Kim, B.H, Kang, S.Y, Song, H.K. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mechanistic Insights into Caffeine Degradation by the Bacterial N-Demethylase Complex.
J.Mol.Biol., 431, 2019
|
|
2L4X
 
 | | Solution Structure of apo-IscU(WT) | | Descriptor: | Iron-sulfur cluster assembly scaffold protein | | Authors: | Kim, J.H, Tonelli, M, Markley, J.L. | | Deposit date: | 2010-10-19 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure and Determinants of Stability of the Iron-Sulfur Cluster Scaffold Protein IscU from Escherichia coli.
Biochemistry, 51, 2012
|
|
8H1A
 
 | |
8H27
 
 | |
8H26
 
 | |
8H0S
 
 | |
8H0T
 
 | |
8H1B
 
 | | Crystal structure of MnmM from S. aureus complexed with SAM and tRNA anti-codon stem loop (ASL) (1.55 A) | | Descriptor: | RNA (5'-R(*AP*CP*GP*GP*AP*CP*UP*UP*UP*GP*AP*CP*UP*CP*CP*GP*U)-3'), S-ADENOSYLMETHIONINE, SODIUM ION, ... | | Authors: | Kim, J, Cho, G, Lee, J. | | Deposit date: | 2022-10-01 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of a novel 5-aminomethyl-2-thiouridine methyltransferase in tRNA modification.
Nucleic Acids Res., 51, 2023
|
|
