6K1G
 
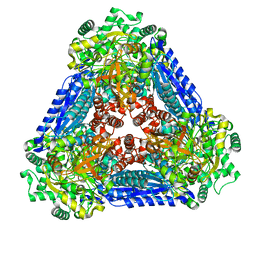 | | Crystal structure of the L-fucose isomerase soaked with Mn2+ from Raoultella sp. | | Descriptor: | L-fucose isomerase, MANGANESE (II) ION | | Authors: | Kim, I.J, Kim, D.H, Nam, K.H, Kim, K.H. | | Deposit date: | 2019-05-10 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Enzymatic synthesis of l-fucose from l-fuculose using a fucose isomerase fromRaoultellasp. and the biochemical and structural analyses of the enzyme.
Biotechnol Biofuels, 12, 2019
|
|
6K1F
 
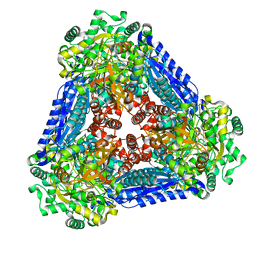 | | Crystal structure of the L-fucose isomerase from Raoultella sp. | | Descriptor: | L-fucose isomerase, MANGANESE (II) ION | | Authors: | Kim, I.J, Kim, D.H, Nam, K.H, Kim, K.H. | | Deposit date: | 2019-05-10 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzymatic synthesis of l-fucose from l-fuculose using a fucose isomerase fromRaoultellasp. and the biochemical and structural analyses of the enzyme.
Biotechnol Biofuels, 12, 2019
|
|
3V9A
 
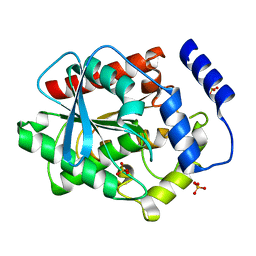 | |
5Y8R
 
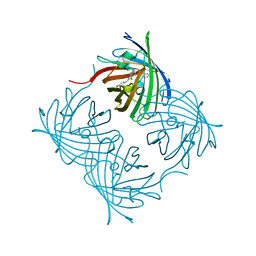 | | ZsYellow at pH 3.5 | | Descriptor: | GFP-like fluorescent chromoprotein FP538 | | Authors: | Bae, J.E, Kim, I.J, Nam, K.H. | | Deposit date: | 2017-08-21 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Disruption of the hydrogen bonding network determines the pH-induced non-fluorescent state of the fluorescent protein ZsYellow by protonation of Glu221.
Biochem. Biophys. Res. Commun., 493, 2017
|
|
5Z6V
 
 | |
5Y8Q
 
 | | ZsYellow at pH 8.0 | | Descriptor: | GFP-like fluorescent chromoprotein FP538 | | Authors: | Bae, J.E, Kim, I.J, Nam, K.H. | | Deposit date: | 2017-08-21 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Disruption of the hydrogen bonding network determines the pH-induced non-fluorescent state of the fluorescent protein ZsYellow by protonation of Glu221.
Biochem. Biophys. Res. Commun., 493, 2017
|
|
5Y4J
 
 | |
5Y4I
 
 | | Crystal structure of glucose isomerase in complex with glycerol in one metal binding mode | | Descriptor: | ACETATE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Bae, J.E, Kim, I.J, Nam, K.H. | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of glucose isomerase in complex with xylitol inhibitor in one metal binding mode
Biochem. Biophys. Res. Commun., 493, 2017
|
|
