6LHV
 
 | | Structure of FANCA and FANCG Complex | | 分子名称: | Fanconi anemia complementation group A, Fanconi anemia complementation group G | | 著者 | Jeong, E, Lee, S, Shin, J, Kim, Y, Scharer, O, Kim, Y, Kim, H, Cho, Y. | | 登録日 | 2019-12-10 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.59 Å) | | 主引用文献 | Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.
Nucleic Acids Res., 48, 2020
|
|
1S1M
 
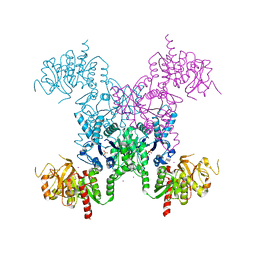 | | Crystal Structure of E. Coli CTP Synthetase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CTP synthase, IODIDE ION, ... | | 著者 | Endrizzi, J.A, Kim, H, Anderson, P.M, Baldwin, E.P. | | 登録日 | 2004-01-06 | | 公開日 | 2004-06-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structure of Escherichia coli Cytidine Triphosphate Synthetase, a Nucleotide-Regulated Glutamine Amidotransferase/ATP-Dependent Amidoligase Fusion Protein and Homologue of Anticancer and Antiparasitic Drug Targets
Biochemistry, 43, 2004
|
|
4O6G
 
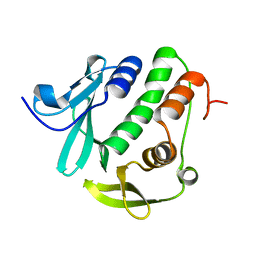 | | Rv3902c from M. tuberculosis | | 分子名称: | Uncharacterized protein | | 著者 | Reddy, B.G, Moates, D.B, Kim, H, Green, T.J, Kim, C, Terwilliger, T.J, Delucas, L.J, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2013-12-20 | | 公開日 | 2014-03-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | 1.55 angstrom resolution X-ray crystal structure of Rv3902c from Mycobacterium tuberculosis.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4WXR
 
 | |
6KF9
 
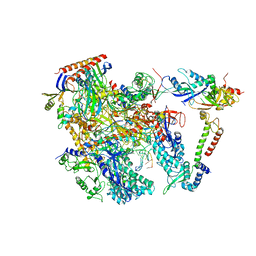 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | 分子名称: | DNA (27-MER), DNA (5'-D(P*TP*CP*GP*GP*TP*AP*AP*TP*CP*AP*CP*GP*CP*TP*CP*C)-3'), DNA-directed RNA polymerase subunit, ... | | 著者 | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | 登録日 | 2019-07-07 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.79 Å) | | 主引用文献 | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
6KF3
 
 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | 分子名称: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit A'', DNA-directed RNA polymerase subunit D, ... | | 著者 | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | 登録日 | 2019-07-06 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
6KF4
 
 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | 分子名称: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit A'', DNA-directed RNA polymerase subunit D, ... | | 著者 | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | 登録日 | 2019-07-06 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.97 Å) | | 主引用文献 | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
5XD0
 
 | | Apo Structure of Beta-1,3-1,4-glucanase from Paenibacillus sp.X4 | | 分子名称: | DI(HYDROXYETHYL)ETHER, Glucanase, TRIETHYLENE GLYCOL | | 著者 | Baek, S.C, Ho, T.-H, Kang, L.-W, Kim, H. | | 登録日 | 2017-03-24 | | 公開日 | 2017-04-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Improvement of enzyme activity of beta-1,3-1,4-glucanase from Paenibacillus sp. X4 by error-prone PCR and structural insights of mutated residues.
Appl. Microbiol. Biotechnol., 101, 2017
|
|
6AF2
 
 | | Crystal structure of N-terminus deletion mutant of Mycobacterium avium diadenosine 5',5'''-P1,P4-tetraphosphate phosphorylase | | 分子名称: | DI(HYDROXYETHYL)ETHER, HIT domain-containing protein, PENTAETHYLENE GLYCOL, ... | | 著者 | Mori, S, Honda, N, Kim, H, Rimbara, E, Shibayama, K. | | 登録日 | 2018-08-08 | | 公開日 | 2019-08-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.001 Å) | | 主引用文献 | Crystal structure of N-terminus deletion mutant of Mycobacterium avium diadenosine tetraphosphate phosphorylase
To Be Published
|
|
7FDA
 
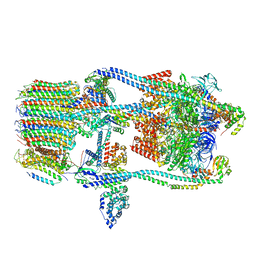 | | CryoEM Structure of Reconstituted V-ATPase, state1 | | 分子名称: | Fusion of yeast V-type proton ATPase subunit H(NT) and human V-type proton ATPase subunit H(CT), V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | 著者 | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | 登録日 | 2021-07-16 | | 公開日 | 2021-12-22 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
7FDB
 
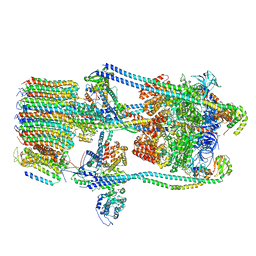 | | CryoEM Structures of Reconstituted V-ATPase,State2 | | 分子名称: | Fusion of yeast V-type proton ATPase subunit H(NT) and human V-type proton ATPase subunit H(CT), V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | 著者 | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | 登録日 | 2021-07-16 | | 公開日 | 2021-12-22 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
7FDC
 
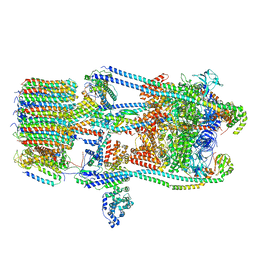 | | CryoEM Structures of Reconstituted V-ATPase, state3 | | 分子名称: | Fusion of yeast V-type proton ATPase subunit H(NT) and human V-type proton ATPase subunit H(CT), V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | 著者 | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | 登録日 | 2021-07-16 | | 公開日 | 2021-12-22 | | 実験手法 | ELECTRON MICROSCOPY (6.6 Å) | | 主引用文献 | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
7FDE
 
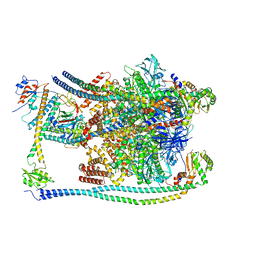 | | CryoEM Structures of Reconstituted V-ATPase, Oxr1 bound V1 | | 分子名称: | Oxidation resistance protein 1, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | 著者 | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | 登録日 | 2021-07-16 | | 公開日 | 2021-12-29 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
2K21
 
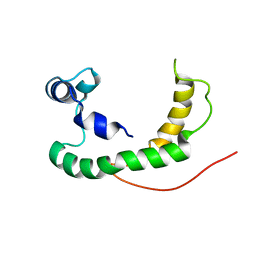 | | NMR structure of human KCNE1 in LMPG micelles at pH 6.0 and 40 degree C | | 分子名称: | Potassium voltage-gated channel subfamily E member | | 著者 | Kang, C, Tian, C, Sonnichsen, F.D, Smith, J.A, Meiler, J, George, A.L, Vanoye, C.G, Sanders, C.R, Kim, H. | | 登録日 | 2008-03-19 | | 公開日 | 2008-12-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of KCNE1 and implications for how it modulates the KCNQ1 potassium channel.
Biochemistry, 47, 2008
|
|
2KDC
 
 | | NMR Solution Structure of E. coli diacylglycerol kinase (DAGK) in DPC micelles | | 分子名称: | Diacylglycerol kinase | | 著者 | Van Horn, W.D, Kim, H, Ellis, C.D, Hadziselimovic, A, Sulistijo, E.S, Karra, M.D, Tian, C, Sonnichsen, F.D, Sanders, C.R. | | 登録日 | 2009-01-06 | | 公開日 | 2009-07-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution nuclear magnetic resonance structure of membrane-integral diacylglycerol kinase
Science, 324, 2009
|
|
2LYP
 
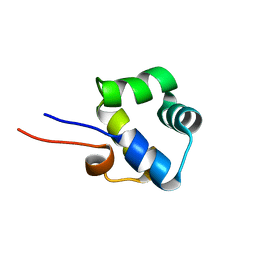 | | NOE-based 3D structure of the monomer of CylR2 in equilibrium with predissociated homodimer at 266K (-7 Celsius degrees) | | 分子名称: | CylR2 | | 著者 | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | 登録日 | 2012-09-19 | | 公開日 | 2013-02-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYQ
 
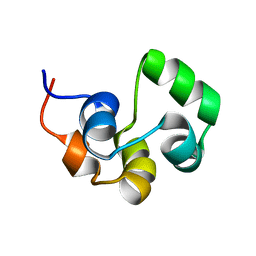 | | NOE-based 3D structure of the monomeric intermediate of CylR2 at 262K (-11 Celsius degrees) | | 分子名称: | CylR2 | | 著者 | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | 登録日 | 2012-09-19 | | 公開日 | 2013-02-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYR
 
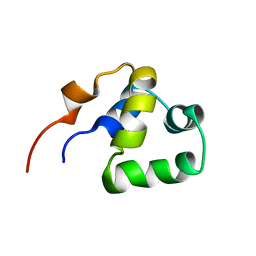 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 259K (-14 Celsius degrees) | | 分子名称: | CylR2 | | 著者 | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | 登録日 | 2012-09-19 | | 公開日 | 2013-02-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYK
 
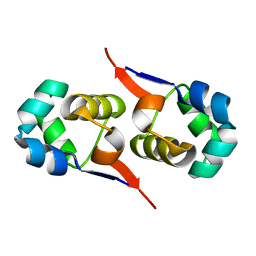 | | NOE-based 3D structure of the CylR2 homodimer at 270K (-3 Celsius degrees) | | 分子名称: | CylR2 | | 著者 | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | 登録日 | 2012-09-19 | | 公開日 | 2013-02-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYL
 
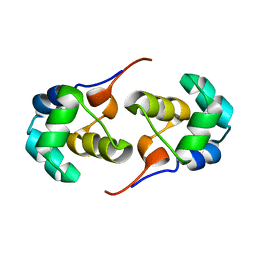 | | NOE-based 3D structure of the predissociated homodimer of CylR2 in equilibrium with monomer at 266K (-7 Celsius degrees) | | 分子名称: | CylR2 | | 著者 | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | 登録日 | 2012-09-19 | | 公開日 | 2013-02-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYS
 
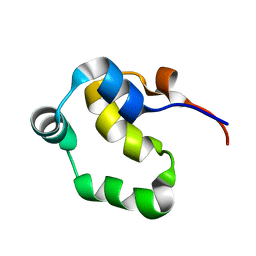 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 257K (-16 Celsius degrees) | | 分子名称: | CylR2 | | 著者 | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | 登録日 | 2012-09-19 | | 公開日 | 2013-02-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYJ
 
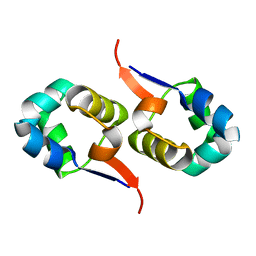 | | NOE-based 3D structure of the CylR2 homodimer at 298K | | 分子名称: | CylR2 | | 著者 | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Giller, K, Becker, S, Zweckstetter, M, Schwieters, C.D. | | 登録日 | 2012-09-19 | | 公開日 | 2013-02-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
6OI5
 
 | | Crystal structure of human Sulfide Quinone Oxidoreductase | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Sulfide:quinone oxidoreductase, mitochondrial | | 著者 | Banerjee, R, Cho, U.S, Kim, H, Moon, S. | | 登録日 | 2019-04-08 | | 公開日 | 2020-01-15 | | 実験手法 | X-RAY DIFFRACTION (2.811 Å) | | 主引用文献 | A Catalytic Trisulfide in Human Sulfide Quinone Oxidoreductase Catalyzes Coenzyme A Persulfide Synthesis and Inhibits Butyrate Oxidation.
Cell Chem Biol, 26, 2019
|
|
6OI6
 
 | | Crystal structure of human Sulfide Quinone Oxidoreductase in complex with coenzyme Q (sulfide soaked) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Sulfide:quinone oxidoreductase, mitochondrial, ... | | 著者 | Banerjee, R, Cho, U.S, Kim, H, Moon, S. | | 登録日 | 2019-04-08 | | 公開日 | 2020-01-15 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | A Catalytic Trisulfide in Human Sulfide Quinone Oxidoreductase Catalyzes Coenzyme A Persulfide Synthesis and Inhibits Butyrate Oxidation.
Cell Chem Biol, 26, 2019
|
|
3SJF
 
 | | X-ray structure of human glutamate carboxypeptidase II in complex with a urea-based inhibitor (A25) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Plechanovova, A, Byun, Y, Alquicer, G, Skultetyova, L, Mlcochova, P, Nemcova, A, Kim, H, Navratil, M, Mease, R, Lubkowski, J, Pomper, M, Konvalinka, J, Rulisek, L, Barinka, C. | | 登録日 | 2011-06-21 | | 公開日 | 2011-10-05 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Novel Substrate-Based Inhibitors of Human Glutamate Carboxypeptidase II with Enhanced Lipophilicity.
J.Med.Chem., 54, 2011
|
|
