1ERH
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE EXTRACELLULAR REGION OF THE COMPLEMENT REGULATORY PROTEIN, CD59, A NEW CELL SURFACE PROTEIN DOMAIN RELATED TO NEUROTOXINS | | Descriptor: | CD59 | | Authors: | Kieffer, B, Driscoll, P.C, Campbell, I.D, Willis, A.C, Van Der Merwe, P.A, Davis, S.J. | | Deposit date: | 1993-12-13 | | Release date: | 1994-04-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the extracellular region of the complement regulatory protein CD59, a new cell-surface protein domain related to snake venom neurotoxins.
Biochemistry, 33, 1994
|
|
1ERG
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE EXTRACELLULAR REGION OF THE COMPLEMENT REGULATORY PROTEIN, CD59, A NEW CELL SURFACE PROTEIN DOMAIN RELATED TO NEUROTOXINS | | Descriptor: | CD59 | | Authors: | Kieffer, B, Driscoll, P.C, Campbell, I.D, Willis, A.C, Van Der Merwe, P.A, Davis, S.J. | | Deposit date: | 1993-12-13 | | Release date: | 1994-04-30 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the extracellular region of the complement regulatory protein CD59, a new cell-surface protein domain related to snake venom neurotoxins.
Biochemistry, 33, 1994
|
|
1Z60
 
 | | Solution structure of the carboxy-terminal domain of human TFIIH P44 subunit | | Descriptor: | TFIIH basal transcription factor complex p44 subunit, ZINC ION | | Authors: | Kellenberger, E, Dominguez, C, Fribourg, S, Wasielewski, E, Moras, D, Poterszman, A, Boelens, R, Kieffer, B. | | Deposit date: | 2005-03-21 | | Release date: | 2005-04-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of TFIIH P44 subunit reveals a novel type of C4C4 ring domain involved in protein-protein interactions.
J.Biol.Chem., 280, 2005
|
|
5IEM
 
 | | NMR structure of the 5'-terminal hairpin of the 7SK snRNA | | Descriptor: | 7SK snRNA | | Authors: | Bourbigot, S, Dock-Bregeon, A.C, Coutant, J, Kieffer, B, Lebars, I. | | Deposit date: | 2016-02-25 | | Release date: | 2016-11-02 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 5'-terminal hairpin of the 7SK small nuclear RNA.
RNA, 22, 2016
|
|
1PFJ
 
 | | Solution structure of the N-terminal PH/PTB domain of the TFIIH P62 subunit | | Descriptor: | TFIIH basal transcription factor complex p62 subunit | | Authors: | Gervais, V, Lamour, V, Jawhari, A, Frindel, F, Wasielewski, E, Thierry, J.C, Kieffer, B, Poterszman, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-05-27 | | Release date: | 2004-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | TFIIH contains a PH domain involved in DNA nucleotide excision repair.
Nat.Struct.Mol.Biol., 11, 2004
|
|
6SQC
 
 | | Crystal structure of complex between nuclear coactivator binding domain of CBP and [1040-1086]ACTR containing alpha-methylated Leu1055 and Leu1076 | | Descriptor: | 1,2-ETHANEDIOL, Maltose/maltodextrin-binding periplasmic protein,CREB-binding protein, Nuclear receptor coactivator 3, ... | | Authors: | Bauer, V, Schmidtgall, B, Gogl, G, Dolenc, j, Osz, J, Kostmann, C, Mitschler, A, Cousido-Siah, A, Rochel, N, Trave, G, Kieffer, B, Torbeev, V. | | Deposit date: | 2019-09-03 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Conformational editing of intrinsically disordered protein by alpha-methylation.
Chem Sci, 12, 2020
|
|
1DW4
 
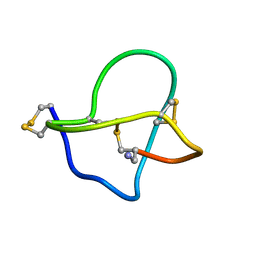 | | NMR STRUCTURE OF OMEGA-CONOTOXIN MVIIA: CONSTRAINTS ON DISULPHIDE BRIDGES | | Descriptor: | OMEGA-CONOTOXIN MVIIA | | Authors: | Atkinson, R.A, Kieffer, B, Dejaegere, A, Sirockin, F, Lefevre, J.-F. | | Deposit date: | 2000-01-24 | | Release date: | 2000-03-01 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic characterization of omega-conotoxin MVIIA: the binding loop exhibits slow conformational exchange.
Biochemistry, 39, 2000
|
|
1DW5
 
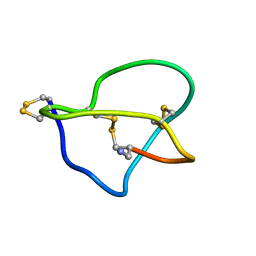 | | NMR STRUCTURE OF OMEGA-CONOTOXIN MVIIA: NO CONSTRAINTS ON DISULPHIDE BRIDGES | | Descriptor: | OMEGA-CONOTOXIN MVIIA | | Authors: | Atkinson, R.A, Kieffer, B, Dejaegere, A, Sirockin, F, Lefevre, J.-F. | | Deposit date: | 2000-01-24 | | Release date: | 2000-03-01 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic characterization of omega-conotoxin MVIIA: the binding loop exhibits slow conformational exchange.
Biochemistry, 39, 2000
|
|
1HFH
 
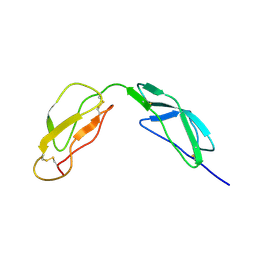 | | SOLUTION STRUCTURE OF A PAIR OF COMPLEMENT MODULES BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | FACTOR H, 15TH AND 16TH C-MODULE PAIR | | Authors: | Barlow, P.N, Steinkasserer, A, Norman, D.G, Kieffer, B, Wiles, A.P, Sim, R.B, Campbell, I.D. | | Deposit date: | 1993-02-23 | | Release date: | 1993-07-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a pair of complement modules by nuclear magnetic resonance.
J.Mol.Biol., 232, 1993
|
|
1HFI
 
 | | SOLUTION STRUCTURE OF A PAIR OF COMPLEMENT MODULES BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | FACTOR H, 15TH C-MODULE PAIR | | Authors: | Barlow, P.N, Steinkasserer, A, Norman, D.G, Kieffer, B, Wiles, A.P, Sim, R.B, Campbell, I.D. | | Deposit date: | 1993-02-23 | | Release date: | 1993-07-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a pair of complement modules by nuclear magnetic resonance.
J.Mol.Biol., 232, 1993
|
|
1G25
 
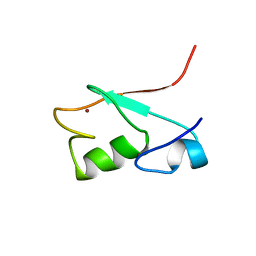 | | SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF THE HUMAN TFIIH MAT1 SUBUNIT | | Descriptor: | CDK-ACTIVATING KINASE ASSEMBLY FACTOR MAT1, ZINC ION | | Authors: | Gervais, V, Wasielewski, E, Busso, D, Poterszman, A, Egly, J.M, Thierry, J.C, Kieffer, B. | | Deposit date: | 2000-10-17 | | Release date: | 2000-11-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the N-terminal Domain of the Human TFIIH MAT1 Subunit: New Insights into the RING Finger Family
J.Biol.Chem., 276, 2001
|
|
5ND9
 
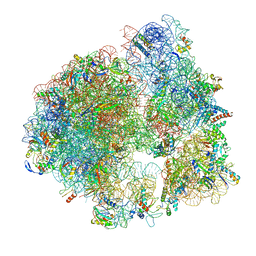 | | Hibernating ribosome from Staphylococcus aureus (Rotated state) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Khusainov, I, Vicens, Q, Ayupov, R, Usachev, K, Myasnikov, A, Simonetti, A, Validov, S, Kieffer, B, Yusupova, G, Yusupov, M, Hashem, Y. | | Deposit date: | 2017-03-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures and dynamics of hibernating ribosomes from Staphylococcus aureus mediated by intermolecular interactions of HPF.
EMBO J., 36, 2017
|
|
5NKO
 
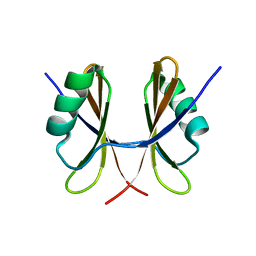 | | Solution structure of the C-terminal domain of S. aureus Hibernating Promoting Factor (CTD-SaHPF) | | Descriptor: | Ribosome hibernation promotion factor | | Authors: | Usachev, K.S, Khusainov, I.S, Ayupov, R.K, Validov, S.Z, Kieffer, B, Yusupov, M.M. | | Deposit date: | 2017-03-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structures and dynamics of hibernating ribosomes from Staphylococcus aureus mediated by intermolecular interactions of HPF.
EMBO J., 36, 2017
|
|
5ND8
 
 | | Hibernating ribosome from Staphylococcus aureus (Unrotated state) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Khusainov, I, Vicens, Q, Ayupov, R, Usachev, K, Myasnikov, A, Simonetti, A, Validov, S, Kieffer, B, Yusupova, G, Yusupov, M, Hashem, Y. | | Deposit date: | 2017-03-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures and dynamics of hibernating ribosomes from Staphylococcus aureus mediated by intermolecular interactions of HPF.
EMBO J., 36, 2017
|
|
2KPK
 
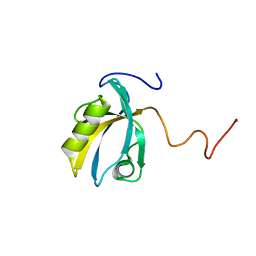 | | MAGI-1 PDZ1 | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 | | Authors: | Charbonnier, S, Nomine, Y, Ramirez, J, Luck, K, Stote, R.H, Trave, G, Kieffer, B, Atkinson, R.A. | | Deposit date: | 2009-10-16 | | Release date: | 2010-10-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The structural and dynamic response of MAGI-1 PDZ1 with non-canonical domain boundaries to binding of human papillomavirus (HPV) E6
J.Mol.Biol., 2011
|
|
2LJX
 
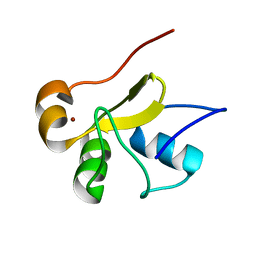 | | Structure of the monomeric N-terminal domain of HPV16 E6 oncoprotein | | Descriptor: | Protein E6, ZINC ION | | Authors: | Zanier, K, Muhamed Sidi, A, Boulade-Ladame, C, Rybin, V, Chappelle, A, Atkinson, A, Kieffer, B, Trave, G. | | Deposit date: | 2011-09-30 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Analysis of the HPV16 E6 Oncoprotein Reveals a Self-Association Mechanism Required for E6-Mediated Degradation of p53.
Structure, 20, 2012
|
|
2LO3
 
 | | Solution structure of Sgf73(59-102) zinc finger domain | | Descriptor: | SAGA-associated factor 73, ZINC ION | | Authors: | Gao, X, Koehler, C, Bonnet, J, Devys, D, Kieffer, B. | | Deposit date: | 2012-01-10 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into the role of SGF11 and SGF73 for the interaction between SAGA and nucleosomes
To be Published
|
|
2JNJ
 
 | | Solution structure of the p8 TFIIH subunit | | Descriptor: | TFIIH basal transcription factor complex TTD-A subunit | | Authors: | Vitorino, M, Atkinson, R.A, Moras, D, Poterszman, A, Kieffer, B, Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2007-01-26 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Self-association Properties of the p8 TFIIH Subunit Responsible for Trichothiodystrophy
J.Mol.Biol., 368, 2007
|
|
2KKT
 
 | | Solution structure of the SCA7 domain of human Ataxin-7-L3 protein | | Descriptor: | Ataxin-7-like protein 3, ZINC ION | | Authors: | Wang, Y, Atkinson, A.R, Bonnet, J, Romier, C, Kieffer, B. | | Deposit date: | 2009-06-29 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Histone deubiquitination by SAGA is modulated by an atypical zinc finger domain of Ataxin-7
To be Published
|
|
2KKR
 
 | | Solution structure of SCA7 zinc finger domain from human ataxin-7 protein | | Descriptor: | Ataxin-7, ZINC ION | | Authors: | Wang, Y, Atkinson, A.R, Bonnet, J, Romier, C, Kieffer, B, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2009-06-29 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Histone deubiquitination by SAGA is modulated by an atypical zinc finger domain of Ataxin-7
To be Published
|
|
2KPL
 
 | | MAGI-1 PDZ1 / E6CT | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1, Protein E6 | | Authors: | Charbonnier, S, Nomine, Y, Ramirez, J, Luck, K, Stote, R.H, Trave, G, Kieffer, B, Atkinson, R.A. | | Deposit date: | 2009-10-16 | | Release date: | 2010-10-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The structural and dynamic response of MAGI-1 PDZ1 with non-canonical domain boundaries to binding of human papillomavirus (HPV) E6
J.Mol.Biol., 2011
|
|
2LJY
 
 | | Haddock model structure of the N-terminal domain dimer of HPV16 E6 | | Descriptor: | Protein E6, ZINC ION | | Authors: | Zanier, K, Muhamed Sidi, A, Boulade-Ladame, C, Rybin, V, Chappelle, A, Atkinson, A, Kieffer, B, Trave, G. | | Deposit date: | 2011-09-30 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Analysis of the HPV16 E6 Oncoprotein Reveals a Self-Association Mechanism Required for E6-Mediated Degradation of p53.
Structure, 20, 2012
|
|
2LO2
 
 | | Solution structure of Sgf11(63-99) zinc finger domain | | Descriptor: | SAGA-associated factor 11, ZINC ION | | Authors: | Gao, X, Koehler, C, Bonnet, J, Devys, D, Kieffer, B. | | Deposit date: | 2012-01-10 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into the role of SGF11 and SGF73 for the interaction between SAGA and nucleosomes
To be Published
|
|
2LJZ
 
 | | Structure of the C-terminal domain of HPV16 E6 oncoprotein | | Descriptor: | Protein E6, ZINC ION | | Authors: | Zanier, K, Muhamed Sidi, A, Boulade-Ladame, C, Rybin, V, Chappelle, A, Atkinson, A, Kieffer, B, Trave, G. | | Deposit date: | 2011-09-30 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Analysis of the HPV16 E6 Oncoprotein Reveals a Self-Association Mechanism Required for E6-Mediated Degradation of p53.
Structure, 20, 2012
|
|
