8JEA
 
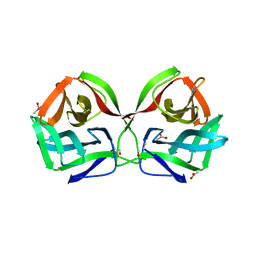 | | Crystal structure of CGL1 from Crassostrea gigas, mannotriose-bound form (CGL1/Man(alpha)1-2Man(alpha)1-2Man) | | Descriptor: | ACETIC ACID, CACODYLATE ION, MAGNESIUM ION, ... | | Authors: | Unno, H, Hatakeyama, T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Mannose oligosaccharide recognition of CGL1, a mannose-specific lectin containing DM9 motifs from Crassostrea gigas, revealed by X-ray crystallographic analysis.
J.Biochem., 175, 2023
|
|
8JEB
 
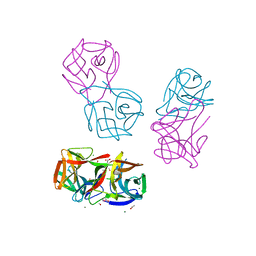 | | Crystal structure of CGL1 from Crassostrea gigas, mannotetraose-bound form (CGL1/Man(alpha)1-2Man(alpha)1-2Man(alpha)1-6Man) | | Descriptor: | ACETIC ACID, MAGNESIUM ION, Natterin-3, ... | | Authors: | Unno, H, Hatakeyama, T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mannose oligosaccharide recognition of CGL1, a mannose-specific lectin containing DM9 motifs from Crassostrea gigas, revealed by X-ray crystallographic analysis.
J.Biochem., 175, 2023
|
|
8JE9
 
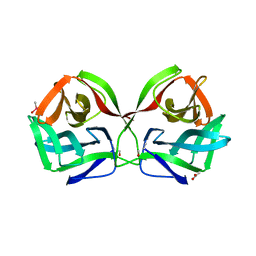 | | Crystal structure of CGL1 from Crassostrea gigas, mannobiose-bound form (CGL1/Man(alpha)1-2Man) | | Descriptor: | ACETIC ACID, CACODYLATE ION, Natterin-3, ... | | Authors: | Unno, H, Hatakeyama, T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Mannose oligosaccharide recognition of CGL1, a mannose-specific lectin containing DM9 motifs from Crassostrea gigas, revealed by X-ray crystallographic analysis.
J.Biochem., 175, 2023
|
|
6TLB
 
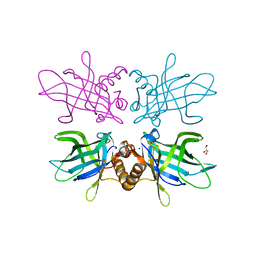 | | Plasmodium falciparum lipocalin (PF3D7_0925900) | | Descriptor: | GLYCEROL, SODIUM ION, Serine/threonine protein kinase | | Authors: | Burda, P.C, Crosskey, T.D, Lauk, K, Wilmanns, M, Gilberger, T.W. | | Deposit date: | 2019-12-02 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | SOLUTION SCATTERING (2.85 Å), X-RAY DIFFRACTION | | Cite: | Structure-Based Identification and Functional Characterization of a Lipocalin in the Malaria Parasite Plasmodium falciparum.
Cell Rep, 31, 2020
|
|
5ZD0
 
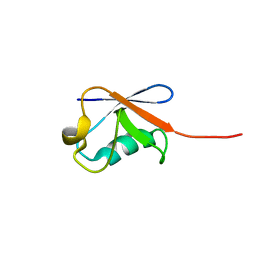 | | Solution structure of human ubiquitin with three alanine mutations in living eukaryotic cells by in-cell NMR spectroscopy | | Descriptor: | ubiquitin | | Authors: | Tanaka, T, Ikeya, T, Kamoshida, H, Mishima, M, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2018-02-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Protein 3D Structure Determination in Living Eukaryotic Cells.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
4WQQ
 
 | | Structure of EPNH mutant of CEL-I | | Descriptor: | CALCIUM ION, Lectin CEL-I, N-acetyl-D-galactosamine-specific C-type, ... | | Authors: | Unno, H, Hatakeyama, T. | | Deposit date: | 2014-10-22 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mannose-recognition mutant of the galactose/N-acetylgalactosamine-specific C-type lectin CEL-I engineered by site-directed mutagenesis.
Biochim.Biophys.Acta, 1850, 2015
|
|
2JZ4
 
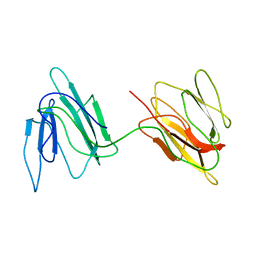 | | Putative 32 kDa myrosinase binding protein At3g16450.1 from Arabidopsis thaliana | | Descriptor: | Jasmonate inducible protein isolog | | Authors: | Takeda, N, Sugimori, N, Torizawa, T, Terauchi, T, Ono, A.M, Yagi, H, Yamaguchi, Y, Kato, K, Ikeya, T, Guntert, P, Aceti, D.J, Markley, J.L, Kainosho, M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-12-28 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of the putative 32 kDa myrosinase-binding protein from Arabidopsis (At3g16450.1) determined by SAIL-NMR.
Febs J., 275, 2008
|
|
5H4S
 
 | |
1VCL
 
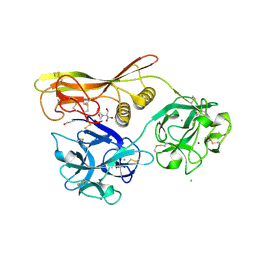 | | Crystal Structure of Hemolytic Lectin CEL-III | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Uchida, T, Yamasaki, T, Eto, S, Sugawara, H, Kurisu, G, Nakagawa, A, Kusunoki, M, Hatakeyama, T. | | Deposit date: | 2004-03-09 | | Release date: | 2004-09-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Hemolytic Lectin CEL-III Isolated from the Marine Invertebrate Cucumaria echinata: IMPLICATIONS OF DOMAIN STRUCTURE FOR ITS MEMBRANE PORE-FORMATION MECHANISM
J.Biol.Chem., 279, 2004
|
|
1WMZ
 
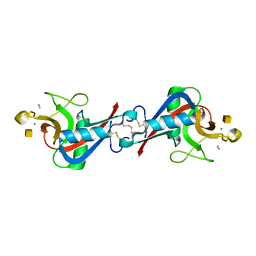 | | Crystal Structure of C-type Lectin CEL-I complexed with N-acetyl-D-galactosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Sugawara, H, Kusunoki, M, Kurisu, G, Fujimoto, T, Aoyagi, H, Hatakeyama, T. | | Deposit date: | 2004-07-22 | | Release date: | 2004-09-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characteristic Recognition of N-Acetylgalactosamine by an Invertebrate C-type Lectin, CEL-I, Revealed by X-ray Crystallographic Analysis
J.Biol.Chem., 279, 2004
|
|
1WMY
 
 | | Crystal Structure of C-type Lectin CEL-I from Cucumaria echinata | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, lectin CEL-I, ... | | Authors: | Sugawara, H, Kusunoki, M, Kurisu, G, Fujimoto, T, Aoyagi, H, Hatakeyama, T. | | Deposit date: | 2004-07-22 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characteristic Recognition of N-Acetylgalactosamine by an Invertebrate C-type Lectin, CEL-I, Revealed by X-ray Crystallographic Analysis
J.Biol.Chem., 279, 2004
|
|
2ROG
 
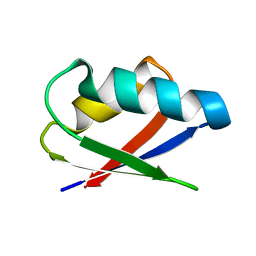 | | Solution structure of Thermus thermophilus HB8 TTHA1718 protein in living E. coli cells | | Descriptor: | Heavy metal binding protein | | Authors: | Sakakibara, D, Sasaki, A, Ikeya, T, Hamatsu, J, Koyama, H, Mishima, M, Mikawa, T, Waelchli, M, Smith, B.O, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2008-03-21 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in living cells by in-cell NMR spectroscopy
Nature, 458, 2009
|
|
2ROE
 
 | | Solution structure of thermus thermophilus HB8 TTHA1718 protein in vitro | | Descriptor: | Heavy metal binding protein | | Authors: | Sakakibara, D, Sasaki, A, Ikeya, T, Hamatsu, J, Koyama, H, Mishima, M, Mikawa, T, Waelchli, M, Smith, B.O, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2008-03-20 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in living cells by in-cell NMR spectroscopy
Nature, 458, 2009
|
|
2JW8
 
 | | Solution structure of stereo-array isotope labelled (SAIL) C-terminal dimerization domain of SARS coronavirus nucleocapsid protein | | Descriptor: | Nucleocapsid protein | | Authors: | Takeda, M, Chang, C, Ikeya, T, Guntert, P, Chang, Y, Hsu, Y, Huang, T, Kainosho, M. | | Deposit date: | 2007-10-06 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the c-terminal dimerization domain of SARS coronavirus nucleocapsid protein solved by the SAIL-NMR method
J.Mol.Biol., 380, 2008
|
|
6M5M
 
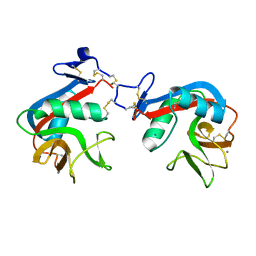 | | SPL-1 - GlcNAc complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, N-acetylglucosamine-specific lectin | | Authors: | Unno, H, Hatakeyama, T. | | Deposit date: | 2020-03-11 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel carbohydrate-recognition mode of the invertebrate C-type lectin SPL-1 from Saxidomus purpuratus revealed by the GlcNAc-complex crystal in the presence of Ca2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
2KBY
 
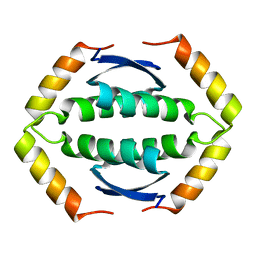 | | The Tetramerization Domain of Human p73 | | Descriptor: | Tumor protein p73 | | Authors: | Coutandin, D, Ikeya, T, Loehr, F, Guntert, P, Ou, H.D, Doetsch, V. | | Deposit date: | 2008-12-12 | | Release date: | 2009-09-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Conformational stability and activity of p73 require a second helix in the tetramerization domain.
Cell Death Differ., 16, 2009
|
|
6A56
 
 | | AJLec from the Sea Anemone Anthopleura japonica | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AJLec, CALCIUM ION, ... | | Authors: | Unno, H, Hatakeyama, T. | | Deposit date: | 2018-06-22 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Identification, Characterization, and X-ray Crystallographic Analysis of a Novel Type of Lectin AJLec from the Sea Anemone Anthopleura japonica.
Sci Rep, 8, 2018
|
|
6A7T
 
 | |
1WN0
 
 | | Crystal Structure of Histidine-containing Phosphotransfer Protein, ZmHP2, from maize | | Descriptor: | histidine-containing phosphotransfer protein | | Authors: | Sugawara, H, Kawano, Y, Hatakeyama, T, Yamaya, T, Kamiya, N, Sakakibara, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-24 | | Release date: | 2005-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the histidine-containing phosphotransfer protein ZmHP2 from maize
Protein Sci., 14, 2005
|
|
6A7S
 
 | | Ca2+-independent C-type lectin SPL-2 from Saxidomus purpuratus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Unno, H, Hatakeyama, T. | | Deposit date: | 2018-07-04 | | Release date: | 2019-03-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel Ca2+-independent carbohydrate recognition of the C-type lectins, SPL-1 and SPL-2, from the bivalve Saxidomus purpuratus.
Protein Sci., 28, 2019
|
|
2M99
 
 | |
3N36
 
 | | Erythrina corallodendron lectin mutant (Y106G) in complex with Galactose | | Descriptor: | CALCIUM ION, Lectin, MANGANESE (II) ION, ... | | Authors: | Thamotharan, S, Karthikeyan, T, Kulkarni, K.A, Shetty, K.N, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2010-05-19 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Modification of the sugar specificity of a plant lectin: structural studies on a point mutant of Erythrina corallodendron lectin.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3N3H
 
 | | Erythrina corallodendron lectin mutant (Y106G) in complex with citrate | | Descriptor: | CALCIUM ION, CITRIC ACID, Lectin, ... | | Authors: | Thamotharan, S, Karthikeyan, T, Kulkarni, K.A, Shetty, K.N, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2010-05-20 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Modification of the sugar specificity of a plant lectin: structural studies on a point mutant of Erythrina corallodendron lectin.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3N35
 
 | | Erythrina corallodendron lectin mutant (Y106G) with N-Acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, CALCIUM ION, Lectin, ... | | Authors: | Thamotharan, S, Karthikeyan, T, Kulkarni, K.A, Shetty, K.N, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2010-05-19 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Modification of the sugar specificity of a plant lectin: structural studies on a point mutant of Erythrina corallodendron lectin.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3W9T
 
 | | pore-forming CEL-III | | Descriptor: | CALCIUM ION, Hemolytic lectin CEL-III, MAGNESIUM ION, ... | | Authors: | Unno, H, Goda, S, Hatakeyama, T. | | Deposit date: | 2013-04-16 | | Release date: | 2014-03-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Hemolytic lectin CEL-III heptamer reveals its transmembrane pore-formation mechanism
J.Biol.Chem., 2014
|
|
