1JNT
 
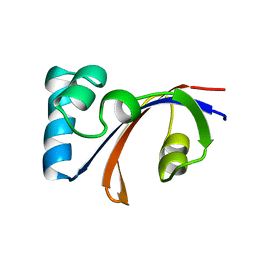 | | NMR Structure of the E. coli Peptidyl-Prolyl cis/trans-Isomerase Parvulin 10 | | Descriptor: | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE C | | Authors: | Kuehlewein, A, Voll, G, Schelbert, B, Kessler, H, Fischer, G, Rahfeld, J.U, Gemmecker, G. | | Deposit date: | 2001-07-25 | | Release date: | 2003-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Escherichia coli Par10: The prototypic member of the Parvulin family of peptidyl-prolyl cis/trans isomerases.
Protein Sci., 13, 2004
|
|
1JNS
 
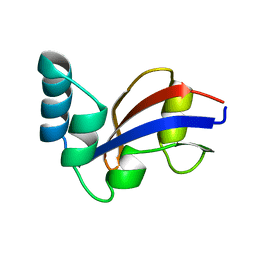 | | NMR Structure of the E. coli Peptidyl-Prolyl cis/trans-Isomerase Parvulin 10 | | Descriptor: | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE C | | Authors: | Kuehlewein, A, Voll, G, Schelbert, B, Kessler, H, Fischer, G, Rahfeld, J.U, Gemmecker, G. | | Deposit date: | 2001-07-25 | | Release date: | 2003-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Escherichia coli Par10: The prototypic member of the Parvulin family of peptidyl-prolyl cis/trans isomerases.
Protein Sci., 13, 2004
|
|
8DRH
 
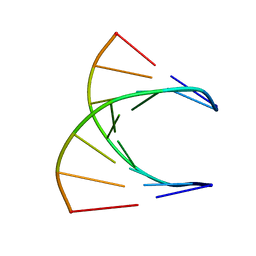 | | HIGH RESOLUTION NMR STRUCTURE OF THE D(GCGTCAGG)R(CCUGACGC) HYBRID, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*CP*AP*GP*G)-3'), RNA (5'-R(*CP*CP*UP*GP*AP*CP*GP*C)-3') | | Authors: | Bachelin, M, Hessler, G, Kurz, G, Hacia, J.G, Dervan, P.B, Kessler, H. | | Deposit date: | 1997-10-13 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a Stereoregular Phosphorothioate DNA/RNA Duplex
Nat.Struct.Biol., 5, 1998
|
|
8PSH
 
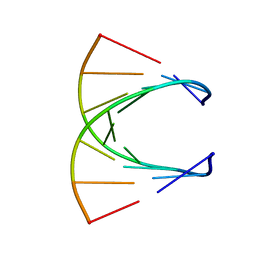 | | HIGH RESOLUTION NMR STRUCTURE OF THE STEREOREGULAR (ALL-RP)-PHOSPHOROTHIOATE-DNA/RNA HYBRID D (G*PS*C*PS*G*PS*T*PS*C*PS*A*PS*G*PS*G)R(CCUGACGC), MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*DGP*(SC)P*(GS)P*(PST)P*(SC)P*(AS)P*(GS)P*(GS))-3'), RNA (5'-R(*CP*CP*UP*GP*AP*CP*GP*C)-3') | | Authors: | Bachelin, M, Hessler, G, Kurz, G, Hacia, J.G, Dervan, P.B, Kessler, H. | | Deposit date: | 1997-10-13 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a Stereoregular Phosphorothioate DNA/RNA Duplex
Nat.Struct.Biol., 5, 1998
|
|
2RMN
 
 | |
3UQ3
 
 | | TPR2AB-domain:pHSP90-complex of yeast Sti1 | | Descriptor: | Heat shock protein, Heat shock protein STI1 | | Authors: | Schmid, A.B, Lagleder, S, Graewert, M.A, Roehl, A, Hagn, F, Wandinger, S.K, Cox, M.B, Demmer, O, Richter, K, Groll, M, Kessler, H, Buchner, J. | | Deposit date: | 2011-11-19 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop.
Embo J., 31, 2012
|
|
3UPV
 
 | | TPR2B-domain:pHsp70-complex of yeast Sti1 | | Descriptor: | Heat shock protein SSA4, Heat shock protein STI1 | | Authors: | Schmid, A.B, Lagleder, S, Graewert, M.A, Roehl, A, Hagn, F, Wandinger, S.K, Cox, M.B, Demmer, O, Richter, K, Groll, M, Kessler, H. | | Deposit date: | 2011-11-18 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop.
Embo J., 31, 2012
|
|
1SVJ
 
 | | The solution structure of the nucleotide binding domain of KdpB | | Descriptor: | Potassium-transporting ATPase B chain | | Authors: | Haupt, M, Bramkamp, M, Coles, M, Altendorf, K, Kessler, H. | | Deposit date: | 2004-03-29 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Inter-domain motions of the N-domain of the KdpFABC complex, a P-type ATPase, are not driven by ATP-induced conformational changes.
J.Mol.Biol., 342, 2004
|
|
1NGL
 
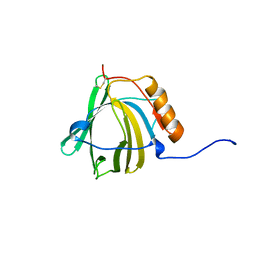 | | HUMAN NEUTROPHIL GELATINASE-ASSOCIATED LIPOCALIN (HNGAL), REGULARISED AVERAGE NMR STRUCTURE | | Descriptor: | PROTEIN (NGAL) | | Authors: | Coles, M, Diercks, T, Muehlenweg, B, Bartsch, S, Zoelzer, V, Tschesche, H, Kessler, H. | | Deposit date: | 1999-02-23 | | Release date: | 1999-05-26 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure and dynamics of human neutrophil gelatinase-associated lipocalin.
J.Mol.Biol., 289, 1999
|
|
1U7Q
 
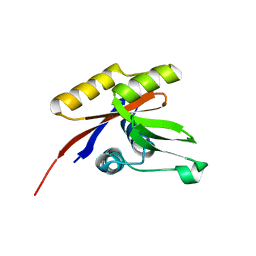 | | THE SOLUTION STRUCTURE OF THE NUCLEOTIDE BINDING DOMAIN OF KDPB | | Descriptor: | Potassium-transporting ATPase B chain | | Authors: | Haupt, M, Bramkamp, M, Coles, M, Altendorf, K, Kessler, H. | | Deposit date: | 2004-08-04 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Inter-domain motions of the N-domain of the KdpFABC complex, a P-type ATPase, are not driven by ATP-induced conformational changes.
J.Mol.Biol., 342, 2004
|
|
2A00
 
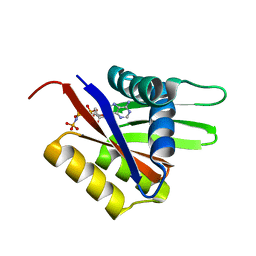 | | The solution structure of the AMP-PNP bound nucleotide binding domain of KdpB | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Potassium-transporting ATPase B chain | | Authors: | Haupt, M, Bramkamp, M, Coles, M, Altendorf, K, Kessler, H. | | Deposit date: | 2005-06-15 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Holo-form of the Nucleotide Binding Domain of the KdpFABC Complex from Escherichia coli Reveals a New Binding Mode
J.Biol.Chem., 281, 2006
|
|
2AKP
 
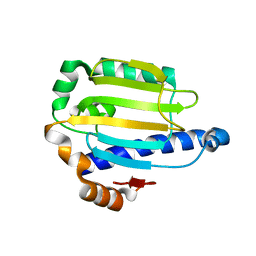 | | Hsp90 Delta24-N210 mutant | | Descriptor: | ATP-dependent molecular chaperone HSP82 | | Authors: | Richter, K, Moser, S, Hagn, F, Friedrich, R, Hainzl, O, Heller, M, Schlee, S, Kessler, H, Reinstein, J, Buchner, J. | | Deposit date: | 2005-08-03 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Intrinsic inhibition of the Hsp90 ATPase activity.
J.Biol.Chem., 281, 2006
|
|
2A29
 
 | | The solution structure of the AMP-PNP bound nucleotide binding domain of KdpB | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Potassium-transporting ATPase B chain | | Authors: | Haupt, M, Bramkamp, M, Coles, M, Altendorf, K, Kessler, H. | | Deposit date: | 2005-06-22 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Holo-form of the Nucleotide Binding Domain of the KdpFABC Complex from Escherichia coli Reveals a New Binding Mode
J.Biol.Chem., 281, 2006
|
|
2CDX
 
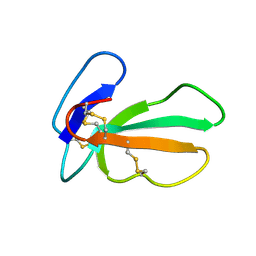 | |
1HZE
 
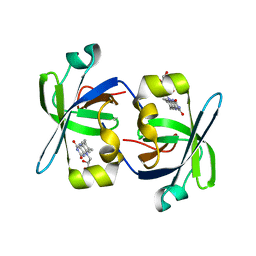 | | SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF RIBOFLAVIN SYNTHASE FROM E. COLI | | Descriptor: | RIBOFLAVIN, RIBOFLAVIN SYNTHASE ALPHA CHAIN | | Authors: | Truffault, V, Coles, M, Diercks, T, Abelmann, K, Eberhardt, S, Luettgen, H, Bacher, A, Kessler, H. | | Deposit date: | 2001-01-24 | | Release date: | 2001-09-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of riboflavin synthase.
J.Mol.Biol., 309, 2001
|
|
1CZ4
 
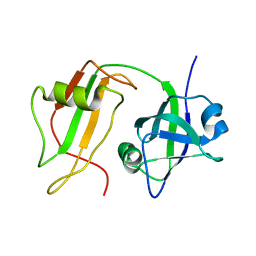 | | NMR STRUCTURE OF VAT-N: THE N-TERMINAL DOMAIN OF VAT (VCP-LIKE ATPASE OF THERMOPLASMA) | | Descriptor: | VCP-LIKE ATPASE | | Authors: | Coles, M, Diercks, T, Liermann, J, Groeger, A, Rockel, B, Baumeister, W, Koretke, K, Lupas, A, Peters, J, Kessler, H. | | Deposit date: | 1999-09-01 | | Release date: | 1999-10-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of VAT-N reveals a 'missing link' in the evolution of complex enzymes from a simple betaalphabetabeta element.
Curr.Biol., 9, 1999
|
|
1CZ5
 
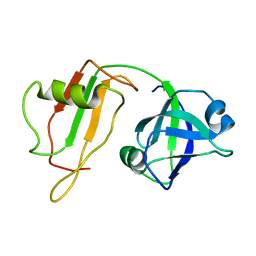 | | NMR STRUCTURE OF VAT-N: THE N-TERMINAL DOMAIN OF VAT (VCP-LIKE ATPASE OF THERMOPLASMA) | | Descriptor: | VCP-LIKE ATPASE | | Authors: | Coles, M, Diercks, T, Liermann, J, Groeger, A, Rockel, B, Baumeister, W, Koretke, K, Lupas, A, Peters, J, Kessler, H. | | Deposit date: | 1999-09-01 | | Release date: | 1999-10-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of VAT-N reveals a 'missing link' in the evolution of complex enzymes from a simple betaalphabetabeta element.
Curr.Biol., 9, 1999
|
|
1IBA
 
 | | GLUCOSE PERMEASE (DOMAIN IIB), NMR, 11 STRUCTURES | | Descriptor: | GLUCOSE PERMEASE | | Authors: | Eberstadt, M, Grdadolnik, S.G, Gemmecker, G, Kessler, H, Buhr, A, Erni, B. | | Deposit date: | 1996-03-23 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the IIB domain of the glucose transporter of Escherichia coli.
Biochemistry, 35, 1996
|
|
1I18
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF RIBOFLAVIN SYNTHASE FROM E. COLI | | Descriptor: | RIBOFLAVIN, RIBOFLAVIN SYNTHASE ALPHA CHAIN | | Authors: | Truffault, V, Coles, M, Diercks, T, Abelmann, K, Eberhardt, S, Luettgen, H, Bacher, A, Kessler, H. | | Deposit date: | 2001-01-31 | | Release date: | 2001-09-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of riboflavin synthase.
J.Mol.Biol., 309, 2001
|
|
2KHM
 
 | | Structure of the C-terminal non-repetitive domain of the spider dragline silk protein ADF-3 | | Descriptor: | Fibroin-3 | | Authors: | Hagn, F.X, Eisoldt, L, Hardy, J.G, Vendrely, C, Coles, M, Scheibel, T, Kessler, H. | | Deposit date: | 2009-04-09 | | Release date: | 2010-04-14 | | Last modified: | 2020-02-26 | | Method: | SOLUTION NMR | | Cite: | A conserved spider silk domain acts as a molecular switch that controls fibre assembly
Nature, 465, 2010
|
|
2LLW
 
 | | Solution structure of the yeast Sti1 DP2 domain | | Descriptor: | Heat shock protein STI1 | | Authors: | Schmid, A.B, Lagleder, S, Graewert, M.A, Roehl, A, Hagn, F, Wandinger, S.K, Cox, M.B, Demmer, O, Richter, K, Groll, M, Kessler, H, Buchner, J. | | Deposit date: | 2011-11-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop.
Embo J., 31, 2012
|
|
2M78
 
 | | [Asp11]RTD-1 | | Descriptor: | [Asp11]RTD-1 | | Authors: | Conibear, A.C, Bochen, A, Rosengren, K, Kessler, H, Craik, D.J. | | Deposit date: | 2013-04-19 | | Release date: | 2014-02-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Cyclic Cystine Ladder of Theta-Defensins as a Stable, Bifunctional Scaffold: A Proof-of-Concept Study Using the Integrin-Binding RGD Motif
Chembiochem, 15, 2014
|
|
2K3G
 
 | |
2M79
 
 | | [Asp2,11]RTD-1 | | Descriptor: | [Asp2,11]RTD-1 | | Authors: | Conibear, A.C, Bochen, A, Rosengren, K, Kessler, H, Craik, D.J. | | Deposit date: | 2013-04-19 | | Release date: | 2014-02-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Cyclic Cystine Ladder of Theta-Defensins as a Stable, Bifunctional Scaffold: A Proof-of-Concept Study Using the Integrin-Binding RGD Motif
Chembiochem, 15, 2014
|
|
2LLV
 
 | | Solution structure of the yeast Sti1 DP1 domain | | Descriptor: | Heat shock protein STI1 | | Authors: | Schmid, A.B, Lagleder, S, Graewert, M.A, Roehl, A, Hagn, F, Wandinger, S.K, Cox, M.B, Demmer, O, Richter, K, Groll, M, Kessler, H, Buchner, J. | | Deposit date: | 2011-11-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop.
Embo J., 31, 2012
|
|
