1SVD
 
 | | The structure of Halothiobacillus neapolitanus RuBisCo | | Descriptor: | GLYCEROL, Ribulose bisphosphate carboxylase small chain, SULFATE ION, ... | | Authors: | Kerfeld, C.A, Sawaya, M.R, Pashkov, I, Cannon, G, Williams, E, Tran, K, Yeates, T.O. | | Deposit date: | 2004-03-29 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of Halothiobacillus neapolitanus RuBisCo
To be Published
|
|
2RCF
 
 | | Carboxysome Shell protein, OrfA from H. Neapolitanus | | Descriptor: | CHLORIDE ION, GLYCEROL, Unidentified carboxysome polypeptide | | Authors: | Kerfeld, C.A, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2007-09-19 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Atomic-level models of the bacterial carboxysome shell.
Science, 319, 2008
|
|
3HIP
 
 | |
1KIB
 
 | | cytochrome c6 from Arthrospira maxima: an assembly of 24 subunits in the form of an oblate shell | | Descriptor: | HEME C, cytochrome c6 | | Authors: | Kerfeld, C.A, Sawaya, M.R, Krogmann, D, Yeates, T.O. | | Deposit date: | 2001-12-03 | | Release date: | 2002-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of cytochrome c6 from Arthrospira maxima: an assembly of 24 subunits in a nearly symmetric shell.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4XB4
 
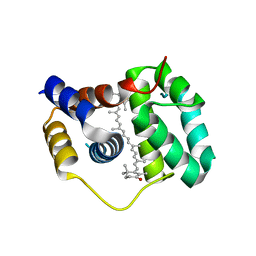 | | Structure of the N-terminal domain of OCP binding canthaxanthin | | Descriptor: | Orange carotenoid-binding protein, beta,beta-carotene-4,4'-dione | | Authors: | Kerfeld, C.A, Sutter, M, Leverenz, R.L. | | Deposit date: | 2014-12-16 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.544 Å) | | Cite: | PHOTOSYNTHESIS. A 12 angstrom carotenoid translocation in a photoswitch associated with cyanobacterial photoprotection.
Science, 348, 2015
|
|
4XB5
 
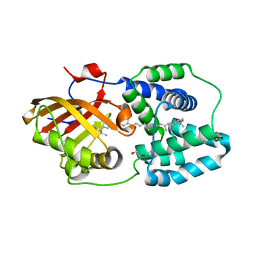 | | Structure of orange carotenoid protein binding canthaxanthin | | Descriptor: | GLYCEROL, Orange carotenoid-binding protein, beta,beta-carotene-4,4'-dione | | Authors: | Kerfeld, C.A, Sutter, M, Leverenz, R.L. | | Deposit date: | 2014-12-16 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | PHOTOSYNTHESIS. A 12 angstrom carotenoid translocation in a photoswitch associated with cyanobacterial photoprotection.
Science, 348, 2015
|
|
5UI2
 
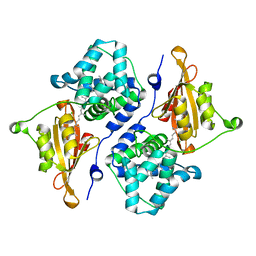 | | CRYSTAL STRUCTURE OF ORANGE CAROTENOID PROTEIN | | Descriptor: | (3'R)-3'-hydroxy-beta,beta-caroten-4-one, CHLORIDE ION, Orange carotenoid-binding protein, ... | | Authors: | KERFELD, C.A, SAWAYA, M.R, VISHNU, B, KROGMANN, D, YEATES, T.O. | | Deposit date: | 2017-01-12 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a cyanobacterial water-soluble carotenoid binding protein.
Structure, 11, 2003
|
|
1MZ4
 
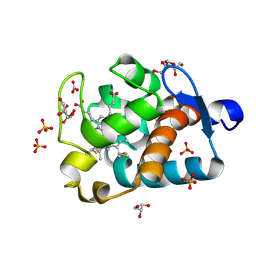 | | Crystal Structure of Cytochrome c550 from Thermosynechococcus elongatus | | Descriptor: | BICARBONATE ION, GLYCEROL, HEME C, ... | | Authors: | Kerfeld, C.A, Sawaya, M.R, Bottin, H, Tran, K.T, Sugiura, M, Kirilovsky, D, Krogmann, D, Yeates, T.O, Boussac, A. | | Deposit date: | 2002-10-05 | | Release date: | 2003-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and EPR characterization of the soluble form of cytochrome c-550 and of the psbV2 gene product from the cyanobacterium Thermosynechococcus elongatus.
Plant Cell.Physiol., 44, 2003
|
|
1CYJ
 
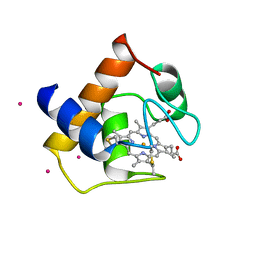 | | CYTOCHROME C6 | | Descriptor: | CADMIUM ION, CYTOCHROME C6, HEME C | | Authors: | Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 1995-05-09 | | Release date: | 1996-01-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of chloroplast cytochrome c6 at 1.9 A resolution: evidence for functional oligomerization.
J.Mol.Biol., 250, 1995
|
|
1CYI
 
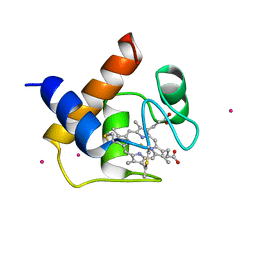 | | CYTOCHROME C6 | | Descriptor: | CADMIUM ION, CYTOCHROME C6, HEME C | | Authors: | Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 1995-05-09 | | Release date: | 1996-01-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of chloroplast cytochrome c6 at 1.9 A resolution: evidence for functional oligomerization.
J.Mol.Biol., 250, 1995
|
|
2A18
 
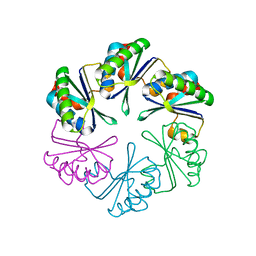 | | carboxysome shell protein ccmK4, crystal form 2 | | Descriptor: | AMMONIUM ION, Carbon dioxide concentrating mechanism protein ccmK homolog 4 | | Authors: | Kerfeld, C.A, Sawaya, M.R, Tanaka, S, Nguyen, C.V, Phillips, M, Beeby, M, Yeates, T.O. | | Deposit date: | 2005-06-18 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Protein structures forming the shell of primitive bacterial organelles
Science, 309, 2005
|
|
1F1F
 
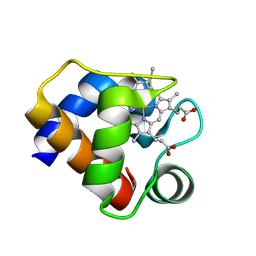 | | CRYSTAL STRUCTURE OF CYTOCHROME C6 FROM ARTHROSPIRA MAXIMA | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Kerfeld, C.A, Serag, A.A, Sawaya, M.R, Krogmann, D.W, Yeates, T.O. | | Deposit date: | 2000-05-18 | | Release date: | 2001-08-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of cytochrome c-549 and cytochrome c6 from the cyanobacterium Arthrospira maxima.
Biochemistry, 40, 2001
|
|
2A1B
 
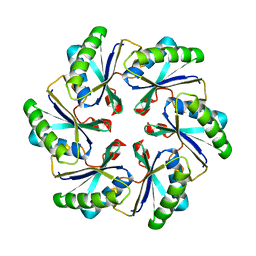 | | Carboxysome shell protein ccmK2 | | Descriptor: | Carbon dioxide concentrating mechanism protein ccmK homolog 2 | | Authors: | Kerfeld, C.A, Sawaya, M.R, Tanaka, S, Nguyen, C.V, Phillips, M, Beeby, M, Yeates, T.O. | | Deposit date: | 2005-06-20 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein structures forming the shell of primitive bacterial organelles
Science, 309, 2005
|
|
2A10
 
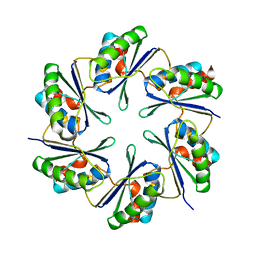 | | carboxysome shell protein ccmK4 | | Descriptor: | Carbon dioxide concentrating mechanism protein ccmK homolog 4 | | Authors: | Kerfeld, C.A, Sawaya, M.R, Tanaka, S, Nguyen, C.V, Phillips, M, Beeby, M, Yeates, T.O. | | Deposit date: | 2005-06-17 | | Release date: | 2005-08-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Protein structures forming the shell of primitive bacterial organelles
Science, 309, 2005
|
|
1F1C
 
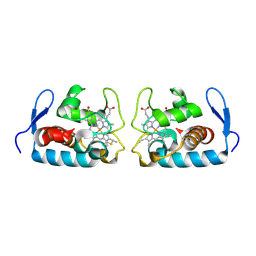 | | CRYSTAL STRUCTURE OF CYTOCHROME C549 | | Descriptor: | CYTOCHROME C549, HEME C | | Authors: | Kerfeld, C.A, Sawaya, M.R, Yeates, T.O, Krogmann, D.W. | | Deposit date: | 2000-05-18 | | Release date: | 2001-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of cytochrome c-549 and cytochrome c6 from the cyanobacterium Arthrospira maxima.
Biochemistry, 40, 2001
|
|
2QW7
 
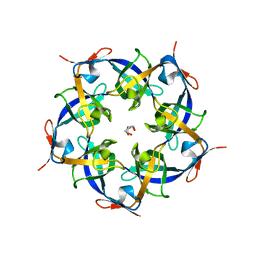 | | Carboxysome Subunit, CcmL | | Descriptor: | Carbon dioxide concentrating mechanism protein ccmL, GLYCEROL | | Authors: | Tanaka, S, Sawaya, M.R, Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 2007-08-09 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atomic-level models of the bacterial carboxysome shell.
Science, 319, 2008
|
|
3U27
 
 | | Crystal structure of ethanolamine utilization protein EutL from Leptotrichia buccalis C-1013-b | | Descriptor: | CALCIUM ION, GLYCEROL, Microcompartments protein, ... | | Authors: | Wu, R, Gu, M, Kerfeld, C.A, Salmeen, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-10-01 | | Release date: | 2012-02-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Crystal structure of ethanolamine utilization protein EutL from Leptotrichia buccalis C-1013-b
To be Published
|
|
5VEG
 
 | | Structure of a Short-Chain Flavodoxin Associated with a Non-Canonical PDU Bacterial Microcompartment | | Descriptor: | CADMIUM ION, FLAVIN MONONUCLEOTIDE, Flavodoxin, ... | | Authors: | Sutter, M, Plegaria, J.S, Kerfeld, C.A. | | Deposit date: | 2017-04-04 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and Functional Characterization of a Short-Chain Flavodoxin Associated with a Noncanonical 1,2-Propanediol Utilization Bacterial Microcompartment.
Biochemistry, 56, 2017
|
|
5C3C
 
 | |
5CUO
 
 | |
5CUP
 
 | |
5DJB
 
 | |
5DIH
 
 | |
5DII
 
 | | Structure of an engineered bacterial microcompartment shell protein binding a [4Fe-4S] cluster | | Descriptor: | IRON/SULFUR CLUSTER, Microcompartments protein | | Authors: | Sutter, M, Aussignargues, C, Turmo, A, Kerfeld, C.A. | | Deposit date: | 2015-09-01 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Structure and Function of a Bacterial Microcompartment Shell Protein Engineered to Bind a [4Fe-4S] Cluster.
J.Am.Chem.Soc., 138, 2016
|
|
5FCY
 
 | | Structure of Anabaena (Nostoc) sp. PCC 7120 Red Carotenoid Protein binding a mixture of carotenoids | | Descriptor: | BETA-CAROTENE, Red carotenoid protein (RCP) | | Authors: | Sutter, M, Leverrenz, R.L, Kerfeld, C.A. | | Deposit date: | 2015-12-15 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.508 Å) | | Cite: | Structure, Diversity, and Evolution of a New Family of Soluble Carotenoid-Binding Proteins in Cyanobacteria.
Mol Plant, 9, 2016
|
|
