5SUX
 
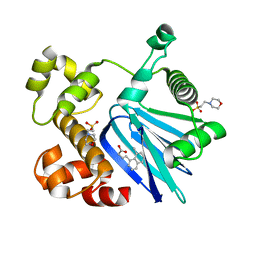 | |
5SUW
 
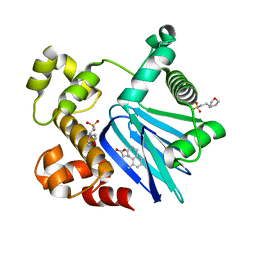 | | Crystal Structure of ToxT from Vibrio Cholerae O395 bound to 3-(8-Methyl-1,2,3,4-tetrahydronaphthalen-1-yl)propanoic acid | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[(1R)-8-methyl-1,2,3,4-tetrahydronaphthalen-1-yl]propanoic acid, TCP pilus virulence regulatory protein | | Authors: | Kull, F.J, Kelley, A.R. | | Deposit date: | 2016-08-04 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of ToxT from Vibrio Cholerae O395 bound to 3-(8-Methyl-1,2,3,4-tetrahydronaphthalen-1-yl)propanoic acid
To Be Published
|
|
4JYA
 
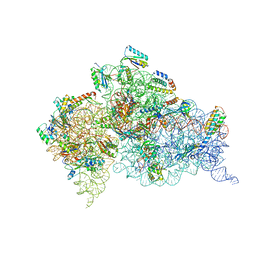 | | Crystal structures of pseudouridinilated stop codons with ASLs | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Fernandez, I.S, Ng, C.L, Kelley, A.C, Guowei, W, Yu, Y.T, Ramakrishnan, V. | | Deposit date: | 2013-03-29 | | Release date: | 2013-06-26 | | Last modified: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Unusual base pairing during the decoding of a stop codon by the ribosome.
Nature, 500, 2013
|
|
4JV5
 
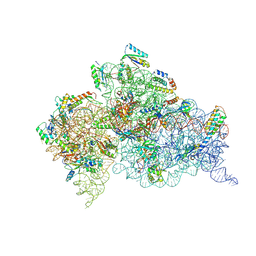 | | Crystal structures of pseudouridinilated stop codons with ASLs | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein 20, 30S ribosomal protein S10, ... | | Authors: | Fernandez, I.S, Ng, C.L, Kelley, A.C, Guowei, W, Yu, Y.T, Ramakrishnan, V. | | Deposit date: | 2013-03-25 | | Release date: | 2013-06-26 | | Last modified: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (3.162 Å) | | Cite: | Unusual base pairing during the decoding of a stop codon by the ribosome.
Nature, 500, 2013
|
|
4KHP
 
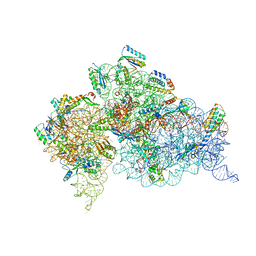 | | Structure of the Thermus thermophilus 30S ribosomal subunit in complex with de-6-MSA-pactamycin | | Descriptor: | 16S Ribosomal RNA, 30S Ribosomal protein S10, 30S Ribosomal protein S11, ... | | Authors: | Tourigny, D.S, Fernandez, I.S, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2013-05-01 | | Release date: | 2013-06-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of a Bioactive Pactamycin Analog Bound to the 30S Ribosomal Subunit.
J.Mol.Biol., 425, 2013
|
|
4K0K
 
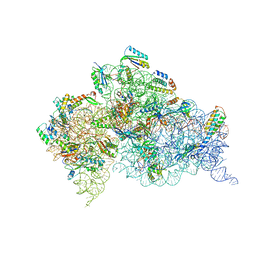 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit complexed with a serine-ASL and mRNA containing a stop codon | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Fernandez, I.S, Ng, C.L, Kelley, A.C, Guowei, W, Yu, Y.T, Ramakrishnan, V. | | Deposit date: | 2013-04-04 | | Release date: | 2013-06-26 | | Last modified: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Unusual base pairing during the decoding of a stop codon by the ribosome.
Nature, 500, 2013
|
|
3IFT
 
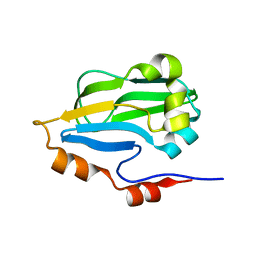 | | Crystal structure of glycine cleavage system protein H from Mycobacterium tuberculosis, using X-rays from the Compact Light Source. | | Descriptor: | Glycine cleavage system H protein | | Authors: | Edwards, T.E, Abendroth, J, Staker, B, Mayer, C, Phan, I, Kelley, A, Analau, E, Leibly, D, Rifkin, J, Loewen, R, Ruth, R.D, Stewart, L.J, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2009-07-25 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure determination of the glycine cleavage system protein H of Mycobacterium tuberculosis using an inverse Compton synchrotron X-ray source.
J.Struct.Funct.Genom., 11, 2010
|
|
