5W0T
 
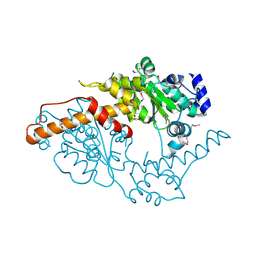 | |
2FFH
 
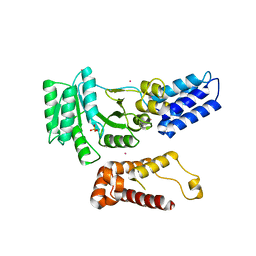 | | THE SIGNAL SEQUENCE BINDING PROTEIN FFH FROM THERMUS AQUATICUS | | Descriptor: | CADMIUM ION, PROTEIN (FFH), SULFATE ION | | Authors: | Keenan, R.J, Freymann, D.M, Walter, P, Stroud, R.M. | | Deposit date: | 1999-06-29 | | Release date: | 1999-07-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the signal sequence binding subunit of the signal recognition particle.
Cell(Cambridge,Mass.), 94, 1998
|
|
2BSW
 
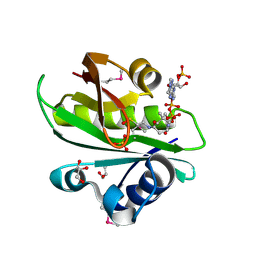 | | Crystal structure of a glyphosate-N-acetyltransferase obtained by DNA shuffling. | | Descriptor: | GLYCEROL, GLYPHOSATE N-ACETYLTRANSFERASE, OXIDIZED COENZYME A, ... | | Authors: | Keenan, R.J, Siehl, D.L, Gorton, R, Castle, L.A. | | Deposit date: | 2005-05-24 | | Release date: | 2005-06-08 | | Last modified: | 2015-10-14 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | DNA Shuffling as a Tool for Protein Crystallization.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
6W6L
 
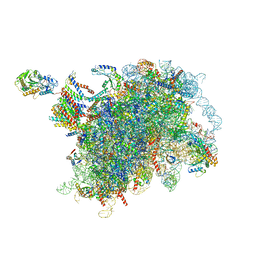 | |
7TM3
 
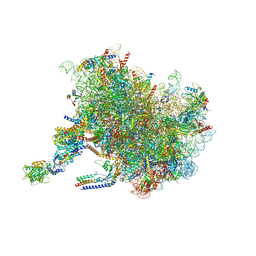 | | Structure of the rabbit 80S ribosome stalled on a 2-TMD Rhodopsin intermediate in complex with the multipass translocon | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Kim, M.K, Lewis, A.J.O, Keenan, R.J, Hegde, R.S. | | Deposit date: | 2022-01-19 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Mechanism of an intramembrane chaperone for multipass membrane proteins.
Nature, 611, 2022
|
|
7TUT
 
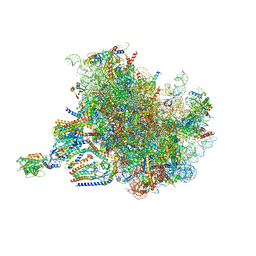 | | Structure of the rabbit 80S ribosome stalled on a 4-TMD Rhodopsin intermediate in complex with the multipass translocon | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Kim, M.K, Lewis, A.J.O, Keenan, R.J, Hegde, R.S. | | Deposit date: | 2022-02-03 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Mechanism of an intramembrane chaperone for multipass membrane proteins.
Nature, 611, 2022
|
|
5C8J
 
 | | A YidC-like protein in the archaeal plasma membrane | | Descriptor: | Antibody fragment, heavy chain, light chain, ... | | Authors: | Borowska, M.T, Dominik, P.K, Anghel, S.A, Kossiakoff, A.A, Keenan, R.J. | | Deposit date: | 2015-06-25 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | A YidC-like Protein in the Archaeal Plasma Membrane.
Structure, 23, 2015
|
|
2WOO
 
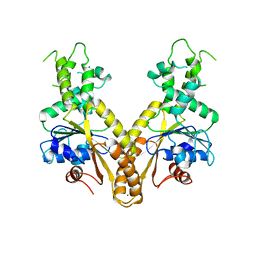 | | Nucleotide-free form of S. pombe Get3 | | Descriptor: | ATPASE GET3, ZINC ION | | Authors: | Mateja, A, Szlachcic, A, Downing, M.E, Dobosz, M, Mariappan, M, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2009-07-27 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.006 Å) | | Cite: | The Structural Basis of Tail-Anchored Membrane Protein Recognition by Get3.
Nature, 461, 2009
|
|
2WOJ
 
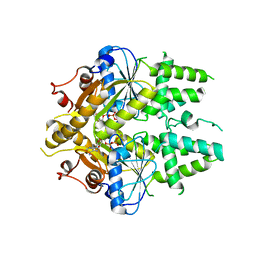 | | ADP-AlF4 complex of S. cerevisiae GET3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPASE GET3, MAGNESIUM ION, ... | | Authors: | Mateja, A, Szlachcic, A, Downing, M.E, Dobosz, M, Mariappan, M, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2009-07-26 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | The Structural Basis of Tail-Anchored Membrane Protein Recognition by Get3.
Nature, 461, 2009
|
|
4XTR
 
 | | Structure of Get3 bound to the transmembrane domain of Pep12 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, ... | | Authors: | Mateja, A, Paduch, M, Chang, H.-Y, Szydlowska, A, Kossiakoff, A.A, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2015-01-23 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo.
Science, 347, 2015
|
|
4XVU
 
 | | Structure of Get3 bound to the transmembrane domain of Nyv1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, Antibody heavy chain, ... | | Authors: | Mateja, A, Paduch, M, Chang, H.-Y, Szydlowska, A, Kossiakoff, A.A, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2015-01-28 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo.
Science, 347, 2015
|
|
4XWO
 
 | | Structure of Get3 bound to the transmembrane domain of Sec22 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, ... | | Authors: | Mateja, A, Paduch, M, Chang, H.-Y, Szydlowska, A, Kossiakoff, A.A, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2015-01-29 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo.
Science, 347, 2015
|
|
3ZQ6
 
 | | ADP-ALF4 COMPLEX OF M. THERM. TRC40 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Sherrill, J, Mariappan, M, Dominik, P, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2011-06-08 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.107 Å) | | Cite: | A Conserved Archaeal Pathway for Tail-Anchored Membrane Protein Insertion.
Traffic, 12, 2011
|
|
3ZS8
 
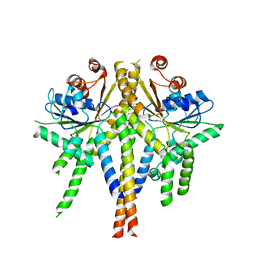 | | S. cerevisiae Get3 complexed with a cytosolic Get1 fragment | | Descriptor: | ATPASE GET3, GOLGI TO ER TRAFFIC PROTEIN 1, ZINC ION | | Authors: | Mariappan, M, Mateja, A, Dobosz, M, Bove, E, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2011-06-24 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Mechanism of Membrane-Associated Steps in Tail-Anchored Protein Insertion.
Nature, 477, 2011
|
|
3ZS9
 
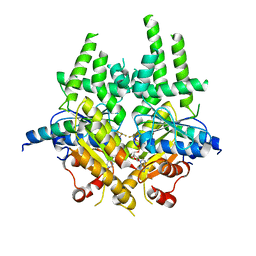 | | S. cerevisiae Get3-ADP-AlF4- complex with a cytosolic Get2 fragment | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPASE GET3, GOLGI TO ER TRAFFIC PROTEIN 2, ... | | Authors: | Mariappan, M, Mateja, A, Dobosz, M, Bove, E, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2011-06-24 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | The Mechanism of Membrane-Associated Steps in Tail-Anchored Protein Insertion.
Nature, 477, 2011
|
|
2JDD
 
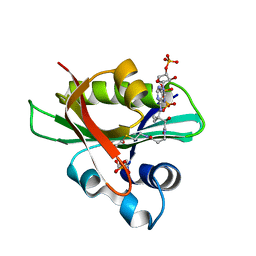 | | Glyphosate N-acetyltransferase bound to acetyl COA and 3-phosphoglycerate | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ACETYL COENZYME *A, GLYPHOSATE N-ACETYLTRANSFERASE, ... | | Authors: | Siehl, D.L, Castle, L.A, Gorton, R, Keenan, R.J. | | Deposit date: | 2007-01-06 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The molecular basis of glyphosate resistance by an optimized microbial acetyltransferase.
J. Biol. Chem., 282, 2007
|
|
2JDC
 
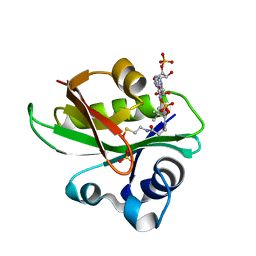 | | Glyphosate N-acetyltransferase bound to oxidized COA and sulfate | | Descriptor: | GLYPHOSATE N-ACETYLTRANSFERASE, OXIDIZED COENZYME A, SULFATE ION | | Authors: | Siehl, D.L, Castle, L.A, Gorton, R, Keenan, R.J. | | Deposit date: | 2007-01-05 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Molecular Basis of Glyphosate Resistance by an Optimized Microbial Acetyltransferase.
J.Biol.Chem., 282, 2007
|
|
2AID
 
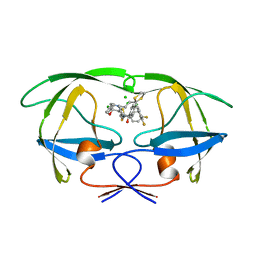 | | STRUCTURE OF A NON-PEPTIDE INHIBITOR COMPLEXED WITH HIV-1 PROTEASE: DEVELOPING A CYCLE OF STRUCTURE-BASED DRUG DESIGN | | Descriptor: | 4-(4-CHLORO-PHENYL)-1-{3-[2-(4-FLUORO-PHENYL)-[1,3]DITHIOLAN-2-YL]-PROPYL}-PIPERIDIN-4-OL, CHLORIDE ION, HUMAN IMMUNODEFICIENCY VIRUS PROTEASE | | Authors: | Rutenber, E.E, Fauman, E.B, Keenan, R.J, Stroud, R.M. | | Deposit date: | 1997-04-17 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a non-peptide inhibitor complexed with HIV-1 protease. Developing a cycle of structure-based drug design.
J.Biol.Chem., 268, 1993
|
|
2VAE
 
 | | Fast maturing red fluorescent protein, DsRed.T4 | | Descriptor: | 1,2-ETHANEDIOL, RED FLUORESCENT PROTEIN | | Authors: | Strongin, D.E, Bevis, B, Khuong, N, Downing, M.E, Strack, R.L, Sundaram, K, Glick, B.S, Keenan, R.J. | | Deposit date: | 2007-08-31 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Rearrangements Near the Chromophore Influence the Maturation Speed and Brightness of Dsred Variants.
Protein Eng.Des.Sel., 20, 2007
|
|
2VAD
 
 | | Monomeric red fluorescent protein, DsRed.M1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, RED FLUORESCENT PROTEIN, ... | | Authors: | Strongin, D.E, Bevis, B, Khuong, N, Downing, M.E, Strack, R.L, Sundaram, K, Glick, B.S, Keenan, R.J. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Rearrangements Near the Chromophore Influence the Maturation Speed and Brightness of Dsred Variants.
Protein Eng.Des.Sel., 20, 2007
|
|
2V4E
 
 | | A non-cytotoxic DsRed variant for whole-cell labeling | | Descriptor: | RED FLUORESCENT PROTEIN DRFP583 | | Authors: | Strack, R.L, Strongin, D.E, Bhattacharyya, D, Tao, W, Berman, A, Broxmeyer, H.E, Keenan, R.J, Glick, B.S. | | Deposit date: | 2008-09-20 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Noncytotoxic Dsred Variant for Whole-Cell Labeling.
Nat.Methods, 5, 2008
|
|
1FFH
 
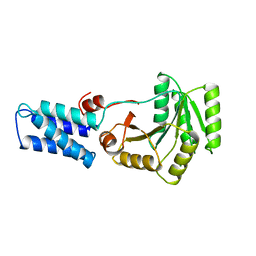 | | N AND GTPASE DOMAINS OF THE SIGNAL SEQUENCE RECOGNITION PROTEIN FFH FROM THERMUS AQUATICUS | | Descriptor: | FFH, MAGNESIUM ION | | Authors: | Freymann, D.M, Keenan, R.J, Stroud, R.M, Walter, P. | | Deposit date: | 1996-12-30 | | Release date: | 1997-12-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the conserved GTPase domain of the signal recognition particle.
Nature, 385, 1997
|
|
1AID
 
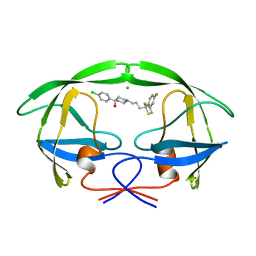 | | STRUCTURE OF A NON-PEPTIDE INHIBITOR COMPLEXED WITH HIV-1 PROTEASE: DEVELOPING A CYCLE OF STRUCTURE-BASED DRUG DESIGN | | Descriptor: | 4-(4-CHLORO-PHENYL)-1-{3-[2-(4-FLUORO-PHENYL)-[1,3]DITHIOLAN-2-YL]-PROPYL}-PIPERIDIN-4-OL, CHLORIDE ION, HUMAN IMMUNODEFICIENCY VIRUS PROTEASE | | Authors: | Rutenber, E.E, Fauman, E.B, Keenan, R.J, Stroud, R.M. | | Deposit date: | 1997-04-16 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a non-peptide inhibitor complexed with HIV-1 protease. Developing a cycle of structure-based drug design.
J.Biol.Chem., 268, 1993
|
|
