5GV8
 
 | |
5GV7
 
 | |
1BM1
 
 | | CRYSTAL STRUCTURE OF BACTERIORHODOPSIN IN THE LIGHT-ADAPTED STATE | | 分子名称: | BACTERIORHODOPSIN, PHOSPHORIC ACID 2,3-BIS-(3,7,11,15-TETRAMETHYL-HEXADECYLOXY)-PROPYL ESTER 2-HYDROXO-3-PHOSPHONOOXY-PROPYL ESTER, RETINAL | | 著者 | Sato, H, Takeda, K, Tani, K, Hino, T, Okada, T, Nakasako, M, Kamiya, N, Kouyama, T. | | 登録日 | 1998-07-28 | | 公開日 | 1999-04-27 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Specific lipid-protein interactions in a novel honeycomb lattice structure of bacteriorhodopsin.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
3HRX
 
 | |
8WU5
 
 | | The complex of CAG repeat sequence-specific binding cPIP and dsDNA with A-A mismatch | | 分子名称: | (1^2Z,4^2Z,11^2Z,14^2Z,22^2Z,25^2Z,32^2Z,35^2Z,19R,40R)-1^1,4^1,11^1,14^1,22^1,25^1,32^1,35^1-octamethyl-2,5,9,12,15,20,23,26,30,33,36,41-dodecaoxo-1^1H,4^1H,11^1H,14^1H,22^1H,25^1H,32^1H,35^1H-3,6,10,13,16,21,24,27,31,34,37,42-dodecaaza-1(2,4),11,22,32(4,2)-tetraimidazola-4,14,25,35(4,2)-tetrapyrrolacyclodotetracontaphane-19,40-diaminium, DNA (5'-D(*GP*CP*(CBR)P*GP*AP*GP*CP*AP*GP*CP*AP*CP*GP*GP*C)-3') | | 著者 | Abe, K, Takeda, K, Sugiyama, H. | | 登録日 | 2023-10-20 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural Studies of a Complex of a CAG/CTG Repeat Sequence-Specific Binding Molecule and A-A-Mismatch-Containing DNA.
Jacs Au, 4, 2024
|
|
7ETQ
 
 | | Crystal structure of Pro-Met-Leu-Leu | | 分子名称: | Pro-Met-Leu-Leu | | 著者 | Kurumida, Y, Ikeda, K, Nakamichi, Y, Hirano, A, Kobayashi, K, Saito, Y, Kameda, T. | | 登録日 | 2021-05-13 | | 公開日 | 2022-05-18 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.04 Å) | | 主引用文献 | Crystal structure of Pro-Met-Leu-Leu
To Be Published
|
|
7ETP
 
 | | Crystal structure of Pro-Phe-Leu-Phe | | 分子名称: | Pro-Phe-Leu-Phe | | 著者 | Kurumida, Y, Ikeda, K, Nakamichi, Y, Hirano, A, Kobayashi, K, Saito, Y, Kameda, T. | | 登録日 | 2021-05-13 | | 公開日 | 2022-05-18 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.09488 Å) | | 主引用文献 | Crystal structure of Pro-Phe-Leu-Phe
To Be Published
|
|
7ETN
 
 | | Crystal structure of Pro-Phe-Leu-Ile | | 分子名称: | PRO-PHE-LEU-ILE | | 著者 | Kurumida, Y, Ikeda, K, Nakamichi, Y, Hirano, A, Kobayashi, K, Saito, Y, Kameda, T. | | 登録日 | 2021-05-13 | | 公開日 | 2022-05-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (0.82 Å) | | 主引用文献 | Crystal structure of Pro-Phe-Leu-Ile
To Be Published
|
|
2YYL
 
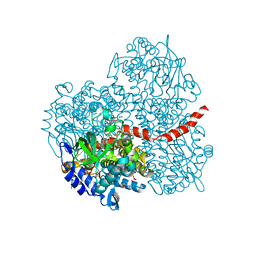 | | Crystal structure of the mutant of HpaB (T198I, A276G, and R466H) complexed with FAD | | 分子名称: | 4-hydroxyphenylacetate-3-hydroxylase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | 著者 | Kim, S.-H, Hisano, T, Takeda, K, Iwasaki, W, Ebihara, A, Miki, K. | | 登録日 | 2007-04-30 | | 公開日 | 2007-09-04 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal Structure of the Oxygenase Component (HpaB) of the 4-Hydroxyphenylacetate 3-Monooxygenase from Thermus thermophilus HB8
J.Biol.Chem., 282, 2007
|
|
2YYG
 
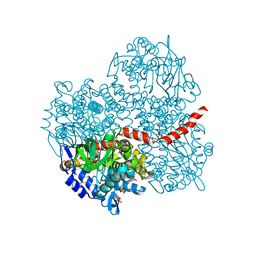 | | Crystal structure of the oxygenase component (HpaB) of 4-hydroxyphenylacetate 3-monooxygenase | | 分子名称: | 4-hydroxyphenylacetate-3-hydroxylase, SULFATE ION | | 著者 | Kim, S.-H, Hisano, T, Takeda, K, Iwasaki, W, Ebihara, A, Miki, K. | | 登録日 | 2007-04-30 | | 公開日 | 2007-09-04 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of the Oxygenase Component (HpaB) of the 4-Hydroxyphenylacetate 3-Monooxygenase from Thermus thermophilus HB8
J.Biol.Chem., 282, 2007
|
|
2YYI
 
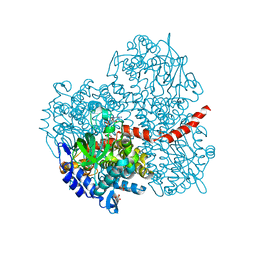 | | Crystal structure of the oxygenase component (HpaB) of 4-hydroxyphenylacetate 3-monooxygenase complexed with FAD | | 分子名称: | 4-hydroxyphenylacetate-3-hydroxylase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | 著者 | Kim, S.-H, Hisano, T, Takeda, K, Iwasaki, W, Ebihara, A, Miki, K. | | 登録日 | 2007-04-30 | | 公開日 | 2007-09-04 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Crystal Structure of the Oxygenase Component (HpaB) of the 4-Hydroxyphenylacetate 3-Monooxygenase from Thermus thermophilus HB8
J.Biol.Chem., 282, 2007
|
|
2YYJ
 
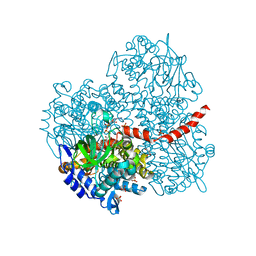 | | Crystal structure of the oxygenase component (HpaB) of 4-hydroxyphenylacetate 3-monooxygenase complexed with FAD and 4-hydroxyphenylacetate | | 分子名称: | 4-HYDROXYPHENYLACETATE, 4-hydroxyphenylacetate-3-hydroxylase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Kim, S.-H, Hisano, T, Takeda, K, Iwasaki, W, Ebihara, A, Miki, K. | | 登録日 | 2007-04-30 | | 公開日 | 2007-09-04 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Crystal Structure of the Oxygenase Component (HpaB) of the 4-Hydroxyphenylacetate 3-Monooxygenase from Thermus thermophilus HB8
J.Biol.Chem., 282, 2007
|
|
2YYM
 
 | | Crystal structure of the mutant of HpaB (T198I, A276G, and R466H) complexed with FAD and 4-hydroxyphenylacetate | | 分子名称: | 4-HYDROXYPHENYLACETATE, 4-hydroxyphenylacetate-3-hydroxylase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Kim, S.-H, Hisano, T, Takeda, K, Iwasaki, W, Ebihara, A, Miki, K. | | 登録日 | 2007-04-30 | | 公開日 | 2007-09-04 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal Structure of the Oxygenase Component (HpaB) of the 4-Hydroxyphenylacetate 3-Monooxygenase from Thermus thermophilus HB8
J.Biol.Chem., 282, 2007
|
|
2YYK
 
 | | Crystal structure of the mutant of HpaB (T198I, A276G, and R466H) | | 分子名称: | 4-hydroxyphenylacetate-3-hydroxylase, ACETIC ACID, GLYCEROL, ... | | 著者 | Kim, S.-H, Hisano, T, Takeda, K, Iwasaki, W, Ebihara, A, Miki, K. | | 登録日 | 2007-04-30 | | 公開日 | 2007-09-04 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structure of the Oxygenase Component (HpaB) of the 4-Hydroxyphenylacetate 3-Monooxygenase from Thermus thermophilus HB8
J.Biol.Chem., 282, 2007
|
|
6KKZ
 
 | | Crystal structure of the S65T/F99S/M153T/V163A variant of perdeuterated GFP at pD 8.5 | | 分子名称: | Green fluorescent protein | | 著者 | Tai, Y, Takaba, K, Hanazono, Y, Dao, H.A, Miki, K, Takeda, K. | | 登録日 | 2019-07-28 | | 公開日 | 2019-12-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | X-ray crystallographic studies on the hydrogen isotope effects of green fluorescent protein at sub-angstrom resolutions
Acta Crystallogr.,Sect.D, 75, 2019
|
|
7VOS
 
 | | High-resolution neutron and X-ray joint refined structure of high-potential iron-sulfur protein in the oxidized state | | 分子名称: | AMMONIUM ION, GLYCEROL, High-potential iron-sulfur protein, ... | | 著者 | Hanazono, Y, Hirano, Y, Takeda, K, Kusaka, K, Tamada, T, Miki, K. | | 登録日 | 2021-10-14 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | NEUTRON DIFFRACTION (0.66 Å), X-RAY DIFFRACTION | | 主引用文献 | Revisiting the concept of peptide bond planarity in an iron-sulfur protein by neutron structure analysis.
Sci Adv, 8, 2022
|
|
3FF5
 
 | | Crystal structure of the conserved N-terminal domain of the peroxisomal matrix-protein-import receptor, Pex14p | | 分子名称: | Peroxisomal biogenesis factor 14, decyl 2-trimethylazaniumylethyl phosphate | | 著者 | Su, J.-R, Takeda, K, Tamura, S, Fujiki, Y, Miki, K. | | 登録日 | 2008-12-01 | | 公開日 | 2008-12-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of the conserved N-terminal domain of the peroxisomal matrix protein import receptor, Pex14p
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3O0T
 
 | | Crystal structure of human phosphoglycerate mutase family member 5 (PGAM5) in complex with phosphate | | 分子名称: | 1,2-ETHANEDIOL, PHOSPHATE ION, Serine/threonine-protein phosphatase PGAM5, ... | | 著者 | Chaikuad, A, Alfano, I, Picaud, S, Filippakopoulos, P, Barr, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Takeda, K, Ichijo, H, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2010-07-20 | | 公開日 | 2010-10-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structures of PGAM5 Provide Insight into Active Site Plasticity and Multimeric Assembly.
Structure, 25, 2017
|
|
3A8I
 
 | | Crystal Structure of ET-EHred-5-CH3-THF complex | | 分子名称: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, Aminomethyltransferase, Glycine cleavage system H protein, ... | | 著者 | Okamura-Ikeda, K, Hosaka, H. | | 登録日 | 2009-10-06 | | 公開日 | 2010-04-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Crystal structure of aminomethyltransferase in complex with dihydrolipoyl-H-protein of the glycine cleavage system: implications for recognition of lipoyl protein substrate, disease-related mutations, and reaction mechanism
J.Biol.Chem., 285, 2010
|
|
3AB9
 
 | | Crystal Structure of lipoylated E. coli H-protein (reduced form) | | 分子名称: | CALCIUM ION, CHLORIDE ION, Glycine cleavage system H protein | | 著者 | Okamura-Ikeda, K, Maita, N. | | 登録日 | 2009-12-04 | | 公開日 | 2010-04-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of aminomethyltransferase in complex with dihydrolipoyl-H-protein of the glycine cleavage system: implications for recognition of lipoyl protein substrate, disease-related mutations, and reaction mechanism
J.Biol.Chem., 285, 2010
|
|
3A8J
 
 | | Crystal Structure of ET-EHred complex | | 分子名称: | Aminomethyltransferase, Glycine cleavage system H protein | | 著者 | Okamura-Ikeda, K, Hosaka, H. | | 登録日 | 2009-10-06 | | 公開日 | 2010-04-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Crystal structure of aminomethyltransferase in complex with dihydrolipoyl-H-protein of the glycine cleavage system: implications for recognition of lipoyl protein substrate, disease-related mutations, and reaction mechanism
J.Biol.Chem., 285, 2010
|
|
7CG3
 
 | | Staggered ring conformation of CtHsp104 (Hsp104 from Chaetomium Thermophilum) | | 分子名称: | Heat shock protein 104 | | 著者 | Inoue, Y, Hanazono, Y, Noi, K, Kawamoto, A, Kimatsuka, M, Harada, R, Takeda, K, Iwamasa, N, Shibata, K, Noguchi, K, Shigeta, Y, Namba, K, Ogura, T, Miki, K, Shinohara, K, Yohda, M. | | 登録日 | 2020-06-30 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Split conformation of Chaetomium thermophilum Hsp104 disaggregase.
Structure, 29, 2021
|
|
6M5B
 
 | | X-ray crystal structure of cyclic-PIP and DNA complex in a reverse binding orientation | | 分子名称: | 1,2-ETHANEDIOL, 4-[[4-[(4-azanyl-1-methyl-pyrrol-2-yl)carbonylamino]-1-methyl-pyrrol-2-yl]carbonylamino]-~{N}-[2-[[(3~{S})-3-azanyl-4-oxidanylidene-butyl]carbamoyl]-1-methyl-imidazol-4-yl]-1-methyl-imidazole-2-carboxamide, DNA (5'-D(*CP*(CBR)P*AP*GP*GP*CP*CP*TP*GP*G)-3'), ... | | 著者 | Abe, K, Hirose, Y, Eki, H, Takeda, K, Bando, T, Endo, M, Sugiyama, H. | | 登録日 | 2020-03-10 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.501 Å) | | 主引用文献 | X-ray Crystal Structure of a Cyclic-PIP-DNA Complex in the Reverse-Binding Orientation.
J.Am.Chem.Soc., 142, 2020
|
|
3MXO
 
 | | Crystal structure oh human phosphoglycerate mutase family member 5 (PGAM5) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Chaikuad, A, Alfano, I, Picaud, S, Filippakopoulos, P, Barr, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Takeda, K, Ichijo, H, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2010-05-07 | | 公開日 | 2010-09-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structures of PGAM5 Provide Insight into Active Site Plasticity and Multimeric Assembly.
Structure, 25, 2017
|
|
3A8K
 
 | | Crystal Structure of ETD97N-EHred complex | | 分子名称: | Aminomethyltransferase, Glycine cleavage system H protein | | 著者 | Okamura-Ikeda, K, Hosaka, H. | | 登録日 | 2009-10-06 | | 公開日 | 2010-04-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of aminomethyltransferase in complex with dihydrolipoyl-H-protein of the glycine cleavage system: implications for recognition of lipoyl protein substrate, disease-related mutations, and reaction mechanism
J.Biol.Chem., 285, 2010
|
|
