6K0Q
 
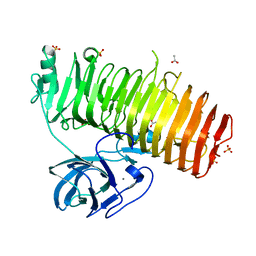 | | Catalytic domain of GH87 alpha-1,3-glucanase D1068A in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.564 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0S
 
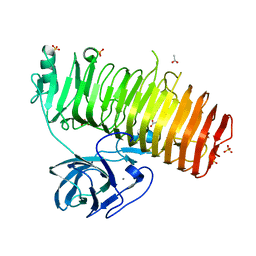 | | Catalytic domain of GH87 alpha-1,3-glucanase D1069A in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0P
 
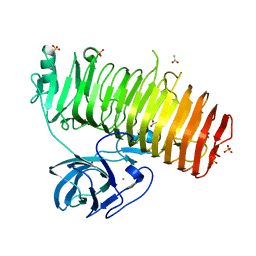 | | Catalytic domain of GH87 alpha-1,3-glucanase D1045A in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.424 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0N
 
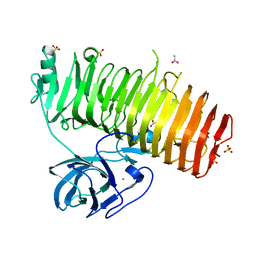 | | Catalytic domain of GH87 alpha-1,3-glucanase in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
8FWS
 
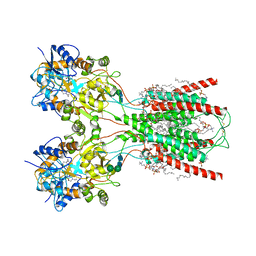 | | Structure of the ligand-binding and transmembrane domains of kainate receptor GluK2 in complex with the positive allosteric modulator BPAM344 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-01-23 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Positive and negative allosteric modulation of GluK2 kainate receptors by BPAM344 and antiepileptic perampanel.
Cell Rep, 42, 2023
|
|
8FWT
 
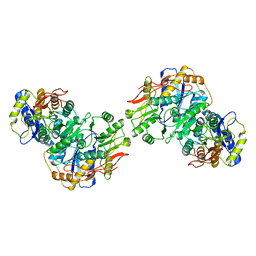 | | Structure of the amino terminal domain of kainate receptor GluK2 in complex with the positive allosteric modulator BPAM344 and competitive antagonist DNQX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Yen, L.Y, Gangwar, S.P, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-01-23 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Positive and negative allosteric modulation of GluK2 kainate receptors by BPAM344 and antiepileptic perampanel.
Cell Rep, 42, 2023
|
|
8FWW
 
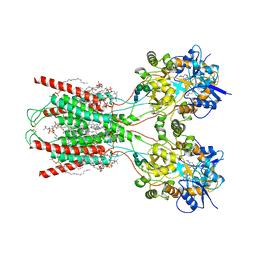 | | Structure of the ligand-binding and transmembrane domains of kainate receptor GluK2 in complex with the positive allosteric modulator BPAM344 and noncompetitive inhibitor perampanel | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-(6'-oxo-1'-phenyl[1',6'-dihydro[2,3'-bipyridine]]-5'-yl)benzonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-01-23 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Positive and negative allosteric modulation of GluK2 kainate receptors by BPAM344 and antiepileptic perampanel.
Cell Rep, 42, 2023
|
|
8FWR
 
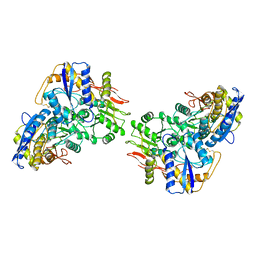 | | Structure of the amino-terminal domain of kainate receptor GluK2 in complex with the positive allosteric modulator BPAM344 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, kainate 2, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-01-23 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Positive and negative allosteric modulation of GluK2 kainate receptors by BPAM344 and antiepileptic perampanel.
Cell Rep, 42, 2023
|
|
8FWV
 
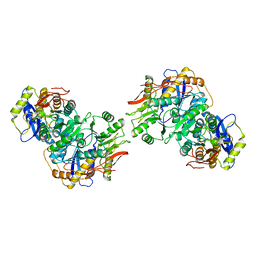 | | Structure of the amino-terminal domain of kainate receptor GluK2 in complex with the positive allosteric modulator BPAM344 and noncompetitive inhibitor perampanel | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-01-23 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Positive and negative allosteric modulation of GluK2 kainate receptors by BPAM344 and antiepileptic perampanel.
Cell Rep, 42, 2023
|
|
8FWU
 
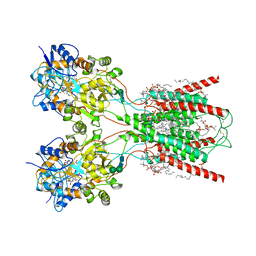 | | Structure of the ligand-binding and transmembrane domains of kainate receptor GluK2 in complex with the positive allosteric modulator BPAM344 and competitive antagonist DNQX | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yen, L.Y, Gangwar, S.P, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-01-23 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Positive and negative allosteric modulation of GluK2 kainate receptors by BPAM344 and antiepileptic perampanel.
Cell Rep, 42, 2023
|
|
6K0V
 
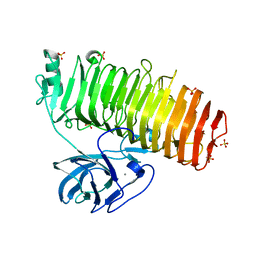 | | Catalytic domain of GH87 alpha-1,3-glucanase D1069A in complex with tetrasaccharides | | Descriptor: | Alpha-1,3-glucanase, CALCIUM ION, SULFATE ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
7C7D
 
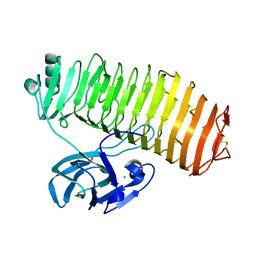 | | Crystal structure of the catalytic unit of thermostable GH87 alpha-1,3-glucanase from Streptomyces thermodiastaticus strain HF3-3 | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, alpha-1,3-glucanase | | Authors: | Itoh, T, Panti, N, Toyotake, Y, Hayashi, J, Suyotha, W, Yano, S, Wakayama, M, Hibi, T. | | Deposit date: | 2020-05-25 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystal structure of the catalytic unit of thermostable GH87 alpha-1,3-glucanase from Streptomyces thermodiastaticus strain HF3-3.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
6DLZ
 
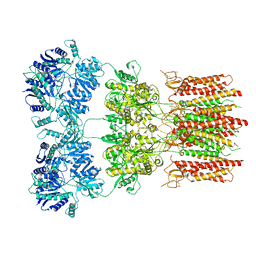 | | Open state GluA2 in complex with STZ after micelle signal subtraction | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Vassilevski, A.A, Sobolevsky, A.I. | | Deposit date: | 2018-06-04 | | Release date: | 2018-08-22 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors.
Neuron, 99, 2018
|
|
7RZA
 
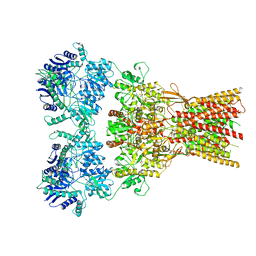 | | Structure of the complex of AMPA receptor GluA2 with auxiliary subunit GSG1L bound to agonist quisqualate | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2 | | Authors: | Gangwar, S.P, Klykov, O.V, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-27 | | Release date: | 2021-10-27 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
7RZ6
 
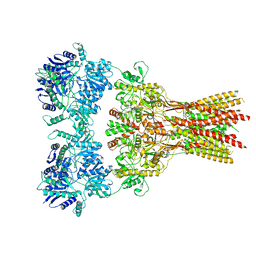 | | Structure of the complex of AMPA receptor GluA2 with auxiliary subunit TARP gamma-5 bound to agonist glutamate | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, GLUTAMIC ACID, Glutamate receptor 2 | | Authors: | Klykov, O.V, Gangwar, S.P, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-27 | | Release date: | 2021-10-27 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
7RYZ
 
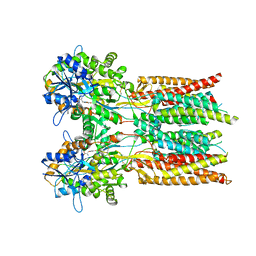 | | Structure of the complex of LBD-TMD part of AMPA receptor GluA2 with auxiliary subunit GSG1L bound to agonist quisqualate | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2 | | Authors: | Gangwar, S.P, Klykov, O.V, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-26 | | Release date: | 2021-10-27 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
7RYY
 
 | | Structure of the complex of LBD-TMD part of AMPA receptor GluA2 with auxiliary subunit TARP gamma-5 bound to agonist glutamate | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, GLUTAMIC ACID, Glutamate receptor 2 | | Authors: | Klykov, O.V, Gangwar, S.P, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-26 | | Release date: | 2021-10-27 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
7RZ9
 
 | |
7RZ7
 
 | | Structure of the complex of AMPA receptor GluA2 with auxiliary subunit TARP gamma-5 bound to agonist Quisqualate | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2 | | Authors: | Klykov, O.V, Gangwar, S.P, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-27 | | Release date: | 2021-10-27 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
5VHW
 
 | | GluA2-0xGSG1L bound to ZK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2,Germ cell-specific gene 1-like protein, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structural Bases of Desensitization in AMPA Receptor-Auxiliary Subunit Complexes.
Neuron, 94, 2017
|
|
5VHZ
 
 | | GluA2-2xGSG1L bound to L-Quisqualate | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2,Germ cell-specific gene 1-like protein | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-03 | | Last modified: | 2017-11-08 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Structural Bases of Desensitization in AMPA Receptor-Auxiliary Subunit Complexes.
Neuron, 94, 2017
|
|
6K0U
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase D1068A in complex with tetrasaccharides | | Descriptor: | Alpha-1,3-glucanase, CALCIUM ION, SULFATE ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
5WEK
 
 | | GluA2 bound to antagonist ZK and GSG1L in digitonin, state 1 | | Descriptor: | Chimera of Glutamate receptor 2,Germ cell-specific gene 1-like protein, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2017-07-10 | | Release date: | 2017-08-02 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Channel opening and gating mechanism in AMPA-subtype glutamate receptors.
Nature, 549, 2017
|
|
5WEO
 
 | | Activated GluA2 complex bound to glutamate, cyclothiazide, and STZ in digitonin | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit chimera | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2017-07-10 | | Release date: | 2017-08-02 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Channel opening and gating mechanism in AMPA-subtype glutamate receptors.
Nature, 549, 2017
|
|
5WEL
 
 | | GluA2 bound to antagonist ZK and GSG1L in digitonin, state 2 | | Descriptor: | Chimera of Glutamate receptor 2, Germ cell-specific gene 1-like protein, Digitonin, ... | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2017-07-10 | | Release date: | 2017-08-02 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Channel opening and gating mechanism in AMPA-subtype glutamate receptors.
Nature, 549, 2017
|
|
