6VFX
 
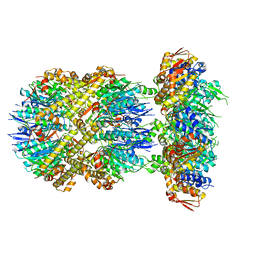 | | ClpXP from Neisseria meningitidis - Conformation B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | Authors: | Ripstein, Z.A, Vahidi, S, Houry, W.A, Rubinstein, J.L, Kay, L.E. | | Deposit date: | 2020-01-06 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A processive rotary mechanism couples substrate unfolding and proteolysis in the ClpXP degradation machinery.
Elife, 9, 2020
|
|
6VFS
 
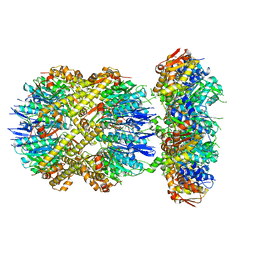 | | ClpXP from Neisseria meningitidis - Conformation A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | Authors: | Ripstein, Z.A, Vahidi, S, Houry, W.A, Rubinstein, J.L, Kay, L.E. | | Deposit date: | 2020-01-06 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A processive rotary mechanism couples substrate unfolding and proteolysis in the ClpXP degradation machinery.
Elife, 9, 2020
|
|
6VGN
 
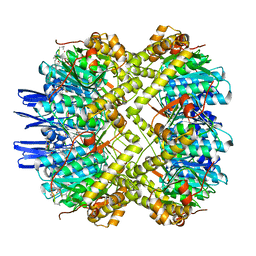 | | ClpP1P2 complex from M. tuberculosis bound to ADEP | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, ATP-dependent Clp protease proteolytic subunit 1, R0M-WFP-ALO-PRO-YCP-ALA-MP8 | | Authors: | Ripstein, Z.A, Vahidi, S, Rubinstein, J.L, Kay, L.E. | | Deposit date: | 2020-01-08 | | Release date: | 2020-03-18 | | Last modified: | 2020-04-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | An allosteric switch regulatesMycobacterium tuberculosisClpP1P2 protease function as established by cryo-EM and methyl-TROSY NMR.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2IL6
 
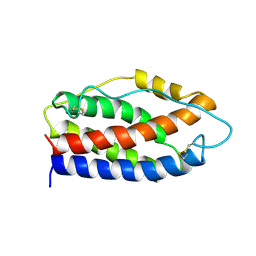 | | HUMAN INTERLEUKIN-6, NMR, 32 STRUCTURES | | Descriptor: | INTERLEUKIN-6 | | Authors: | Xu, G.Y, Yu, H.A, Hong, J, Stahl, M, Mcdonagh, T, Kay, L.E, Cumming, D.A. | | Deposit date: | 1997-01-31 | | Release date: | 1998-02-04 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of recombinant human interleukin-6.
J.Mol.Biol., 268, 1997
|
|
1THQ
 
 | | Crystal Structure of Outer Membrane Enzyme PagP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CrcA protein, ... | | Authors: | Ahn, V.E, Lo, E.I, Engel, C.K, Chen, L, Hwang, P.M, Kay, L.E, Bishop, R.E, Prive, G.G. | | Deposit date: | 2004-06-01 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A hydrocarbon ruler measures palmitate in the enzymatic acylation of endotoxin.
Embo J., 23, 2004
|
|
1TBA
 
 | | SOLUTION STRUCTURE OF A TBP-TAFII230 COMPLEX: PROTEIN MIMICRY OF THE MINOR GROOVE SURFACE OF THE TATA BOX UNWOUND BY TBP, NMR, 25 STRUCTURES | | Descriptor: | TRANSCRIPTION INITIATION FACTOR IID 230K CHAIN, TRANSCRIPTION INITIATION FACTOR TFIID | | Authors: | Liu, D, Ishima, R, Tong, K.I, Bagby, S, Kokubo, T, Muhandiram, D.R, Kay, L.E, Nakatani, Y, Ikura, M. | | Deposit date: | 1998-08-16 | | Release date: | 1999-08-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a TBP-TAF(II)230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP.
Cell(Cambridge,Mass.), 94, 1998
|
|
1IL6
 
 | | HUMAN INTERLEUKIN-6, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | INTERLEUKIN-6 | | Authors: | Xu, G.Y, Yu, H.A, Hong, J, Stahl, M, Mcdonagh, T, Kay, L.E, Cumming, D.A. | | Deposit date: | 1997-01-31 | | Release date: | 1998-02-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of recombinant human interleukin-6.
J.Mol.Biol., 268, 1997
|
|
2K3B
 
 | |
2JSS
 
 | | NMR structure of chaperone Chz1 complexed with histone H2A.Z-H2B | | Descriptor: | Chimera of Histone H2B.1 and Histone H2A.Z, Uncharacterized protein YER030W | | Authors: | Zhou, Z, Feng, H, Hansen, D.F, Kato, H, Luk, E, Freedberg, D.I, Kay, L.E, Wu, C, Bai, Y. | | Deposit date: | 2007-07-11 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of chaperone Chz1 complexed with histones H2A.Z-H2B.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2KU2
 
 | |
2LKS
 
 | | Ff11-60 | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Barette, J, Velyvis, A, Religa, T.L, Korzhnev, D.M, Kay, L.E. | | Deposit date: | 2011-10-19 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cross-Validation of the Structure of a Transiently Formed and Low Populated FF Domain Folding Intermediate Determined by Relaxation Dispersion NMR and CS-Rosetta.
J.Phys.Chem.B, 116, 2012
|
|
2KU1
 
 | |
2L9V
 
 | | NMR structure of the FF domain L24A mutant's folding transition state | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Korzhnev, D.M, Vernon, R.M, Religa, T.L, Hansen, A, Baker, D, Fersht, A.R, Kay, L.E. | | Deposit date: | 2011-02-24 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nonnative interactions in the FF domain folding pathway from an atomic resolution structure of a sparsely populated intermediate: an NMR relaxation dispersion study.
J.Am.Chem.Soc., 133, 2011
|
|
2LCB
 
 | | Solution Structure of a Minor and Transiently Formed State of a T4 Lysozyme Mutant | | Descriptor: | Lysozyme | | Authors: | Bouvignies, G, Vallurupalli, P, Hansen, D, Correia, B, Lange, O, Bah, A, Vernon, R.M, Dahlquist, F.W, Baker, D, Kay, L.E. | | Deposit date: | 2011-04-26 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a minor and transiently formed state of a T4 lysozyme mutant.
Nature, 477, 2011
|
|
2L2P
 
 | | Folding Intermediate of the Fyn SH3 A39V/N53P/V55L from NMR Relaxation Dispersion Experiments | | Descriptor: | Tyrosine-protein kinase Fyn | | Authors: | Neudecker, P, Robustelli, P, Cavalli, A, Vendruscolo, M, Kay, L.E. | | Deposit date: | 2010-08-25 | | Release date: | 2011-09-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of an intermediate state in protein folding and aggregation.
Science, 336, 2012
|
|
2LP5
 
 | | Native Structure of the Fyn SH3 A39V/N53P/V55L | | Descriptor: | Tyrosine-protein kinase Fyn | | Authors: | Neudecker, P, Robustelli, P, Cavalli, A, Vendruscolo, M, Kay, L.E. | | Deposit date: | 2012-02-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of an intermediate state in protein folding and aggregation.
Science, 336, 2012
|
|
2LC9
 
 | | Solution Structure of a Minor and Transiently Formed State of a T4 Lysozyme Mutant | | Descriptor: | Lysozyme | | Authors: | Bouvignies, G, Vallurupalli, P, Hansen, D, Correia, B, Lange, O, Bah, A, Vernon, R.M, Dahlquist, F.W, Baker, D, Kay, L.E. | | Deposit date: | 2011-04-26 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a minor and transiently formed state of a T4 lysozyme mutant.
Nature, 477, 2011
|
|
2KZG
 
 | | A Transient and Low Populated Protein Folding Intermediate at Atomic Resolution | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Korzhnev, D.M, Religa, T.L, Banachewicz, W, Fersht, A.R, Kay, L.E. | | Deposit date: | 2010-06-17 | | Release date: | 2010-09-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A transient and low-populated protein-folding intermediate at atomic resolution.
Science, 329, 2010
|
|
7KR2
 
 | |
6ZL7
 
 | | CRYSTAL STRUCTURE OF C173S MUTATION IN THE PMGL2 ESTERASE FROM PERMAFROST METAGENOMIC LIBRARY | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, MAGNESIUM ION, PMGL2 | | Authors: | Goryaynova, D.A, Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Kryukova, M.V, Petrovskaya, L.E, Novototskaya-Vlasova, K.A, Rivkina, E.M, Dolgikh, D.A, Kirpichnikov, M.P, Popov, V.O. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF C173S MUTATION IN THE PMGL2 ESTERASE FROM PERMAFROST METAGENOMIC LIBRARY
To Be Published
|
|
7B1X
 
 | | Crystal structure of cold-active esterase PMGL3 from permafrost metagenomic library | | Descriptor: | esterase PMGL3 | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Petrovskaya, L.E, Kryukova, M.V, Kryukova, E.A, Korzhenevsky, D.A, Lomakina, G.Y, Novototskaya-Vlasova, K.A, Rivkina, E.M, Dolgikh, D.A, Kirpichnikov, M.P, Popov, V.O. | | Deposit date: | 2020-11-25 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Characterization of a Cold-Active PMGL3 Esterase with Unusual Oligomeric Structure.
Biomolecules, 11, 2021
|
|
6QIN
 
 | | CRYSTAL STRUCTURE OF THE PMGL2 ESTERASE FROM PERMAFROST METAGENOMIC LIBRARY | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PMGL2 | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Kryukova, M.V, Petrovskaya, L.E, Novototskaya-Vlasova, K.A, Rivkina, E.M, Dolgikh, D.A, Kirpichnikov, M.P, Popov, V.O. | | Deposit date: | 2019-01-21 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of PMGL2 esterase from the hormone-sensitive lipase family with GCSAG motif around the catalytic serine.
Plos One, 15, 2020
|
|
6QLA
 
 | | CRYSTAL STRUCTURE OF THE PMGL2 ESTERASE (point mutant 1) FROM PERMAFROST METAGENOMIC LIBRARY | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Boyko, K.M, Garsia, D, Nikolaeva, A.Y, Korzhenevskiy, D.A, Kryukova, M.V, Petrovskaya, L.E, Novototskaya-Vlasova, K.A, Rivkina, E.M, Dolgikh, D.A, Kirpichnikov, M.P, Popov, V.O. | | Deposit date: | 2019-01-31 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of PMGL2 esterase from the hormone-sensitive lipase family with GCSAG motif around the catalytic serine.
Plos One, 15, 2020
|
|
4NS4
 
 | | Crystal structure of cold-active estarase from Psychrobacter cryohalolentis K5T | | Descriptor: | Alpha/beta hydrolase fold protein | | Authors: | Boyko, K.M, Petrovskaya, L.E, Gorbacheva, M.A, Korgenevsky, D.A, Novototskaya-Vlasova, K.A, Rivkina, E.M, Dolgikh, D.A, Kirpichnikov, M.P, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2013-11-28 | | Release date: | 2015-01-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Three-dimentional structure of an esterse from Psychrobacter cryohalolentis K5T provides clues to unusual thermostability of a cold-active enzyme
To be Published
|
|
