8H25
 
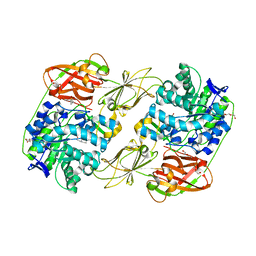 | | Lacticaseibacillus casei GH35 beta-galactosidase LBCZ_0230 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-galactosidase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Saburi, W, Ota, T, Kato, K, Tagami, T, Yamashita, K, Yao, M, Mori, H. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Function and Structure of Lacticaseibacillus casei GH35 beta-Galactosidase LBCZ_0230 with High Hydrolytic Activity to Lacto- N -biose I and Galacto- N -biose.
J Appl Glycosci (1999), 70, 2023
|
|
5AY9
 
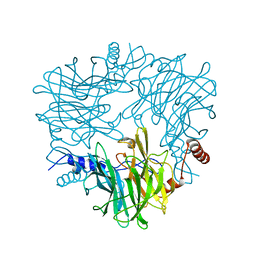 | |
5AYC
 
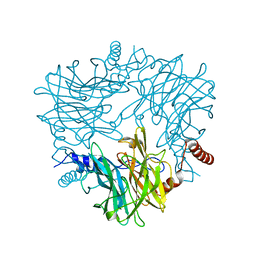 | | Crystal structure of Ruminococcus albus 4-O-beta-D-mannosyl-D-glucose phosphorylase (RaMP1) in complexes with sulfate and 4-O-beta-D-mannosyl-D-glucose | | Descriptor: | 4-O-beta-D-mannosyl-D-glucose phosphorylase, SULFATE ION, beta-D-mannopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Ye, Y, Saburi, W, Kato, K, Yao, M. | | Deposit date: | 2015-08-13 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the difference in substrate recognition of two mannoside phosphorylases from two GH130 subfamilies.
Febs Lett., 590, 2016
|
|
1X60
 
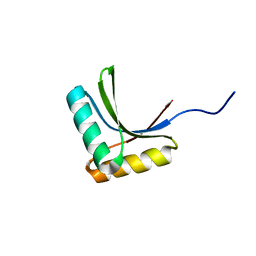 | | Solution structure of the peptidoglycan binding domain of B. subtilis cell wall lytic enzyme CwlC | | Descriptor: | Sporulation-specific N-acetylmuramoyl-L-alanine amidase | | Authors: | Mishima, M, Shida, T, Yabuki, K, Kato, K, Sekiguchi, J, Kojima, C. | | Deposit date: | 2005-05-17 | | Release date: | 2005-08-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Peptidoglycan Binding Domain of Bacillus subtilis Cell Wall Lytic Enzyme CwlC: Characterization of the Sporulation-Related Repeats by NMR(,)
Biochemistry, 44, 2005
|
|
5H18
 
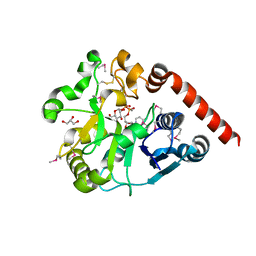 | | Crystal structure of catalytic domain of UGGT (UDP-glucose-bound form) from Thermomyces dupontii | | Descriptor: | CALCIUM ION, GLYCEROL, UGGT, ... | | Authors: | Satoh, T, Zhu, T, Toshimori, T, Kamikubo, H, Uchihashi, T, Kato, K. | | Deposit date: | 2016-10-08 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Visualisation of a flexible modular structure of the ER folding-sensor enzyme UGGT.
Sci Rep, 7, 2017
|
|
1WXS
 
 | | Solution Structure of Ufm1, a ubiquitin-fold modifier | | Descriptor: | Ubiquitin-fold Modifier 1 | | Authors: | Sasakawa, H, Sakata, E, Yamaguchi, Y, Komatsu, M, Tatsumi, K, Kominami, E, Tanaka, K, Kato, K. | | Deposit date: | 2005-02-01 | | Release date: | 2006-04-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of Ufm1, a ubiquitin-fold modifier 1
Biochem.Biophys.Res.Commun., 343, 2006
|
|
5GIJ
 
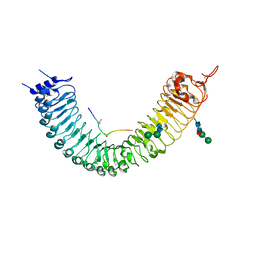 | | Crystal structure of TDR-TDIF complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich repeat receptor-like protein kinase TDR, ... | | Authors: | Morita, J, Kato, K, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2016-06-23 | | Release date: | 2016-08-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the plant receptor-like kinase TDR in complex with the TDIF peptide
Nat Commun, 7, 2016
|
|
4YHD
 
 | | Staphylococcal alpha-hemolysin H35A mutant monomer | | Descriptor: | Alpha-hemolysin, CHLORIDE ION | | Authors: | Sugawara, T, Kato, K, Tanaka, Y, Yao, M. | | Deposit date: | 2015-02-27 | | Release date: | 2015-10-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for pore-forming mechanism of staphylococcal alpha-hemolysin
Toxicon, 108, 2015
|
|
5AXN
 
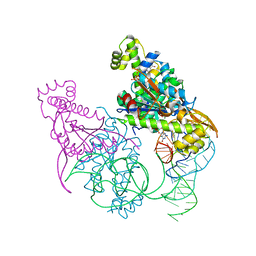 | | Crystal structure of Thg1 like protein (TLP) with tRNA(Phe) and GDPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, RNA (75-MER), ... | | Authors: | Kimura, S, Suzuki, T, Yu, J, Kato, K, Yao, M. | | Deposit date: | 2015-07-31 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Template-dependent nucleotide addition in the reverse (3'-5') direction by Thg1-like protein
Sci Adv, 2, 2016
|
|
2JZ4
 
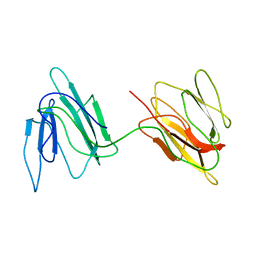 | | Putative 32 kDa myrosinase binding protein At3g16450.1 from Arabidopsis thaliana | | Descriptor: | Jasmonate inducible protein isolog | | Authors: | Takeda, N, Sugimori, N, Torizawa, T, Terauchi, T, Ono, A.M, Yagi, H, Yamaguchi, Y, Kato, K, Ikeya, T, Guntert, P, Aceti, D.J, Markley, J.L, Kainosho, M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-12-28 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of the putative 32 kDa myrosinase-binding protein from Arabidopsis (At3g16450.1) determined by SAIL-NMR.
Febs J., 275, 2008
|
|
1IYF
 
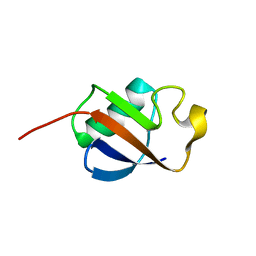 | | Solution structure of ubiquitin-like domain of human parkin | | Descriptor: | parkin | | Authors: | Sakata, E, Yamaguchi, Y, Kurimoto, E, Kikuchi, J, Yokoyama, S, Kawahara, H, Yokosawa, H, Hattori, N, Mizuno, Y, Tanaka, K, Kato, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-08-13 | | Release date: | 2003-03-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Parkin binds the Rpn10 subunit of 26S proteasomes through its ubiquitin-like domain
EMBO REP., 4, 2003
|
|
8IN4
 
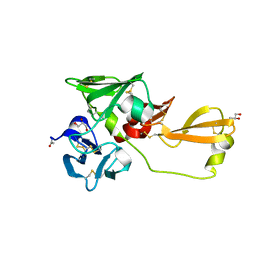 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai | | Descriptor: | 25 kDa polyphenol-binding protein, ACETYL GROUP, GLYCEROL | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN1
 
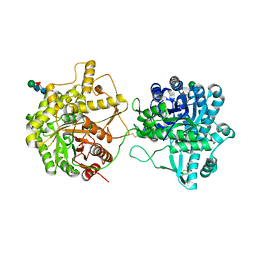 | | beta-glucosidase protein from Aplysia kurodai | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-Glucosidase, alpha-L-fucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN3
 
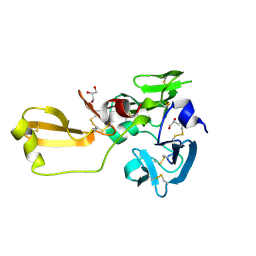 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai | | Descriptor: | 25 kDa polyphenol-binding protein, GLYCEROL | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN6
 
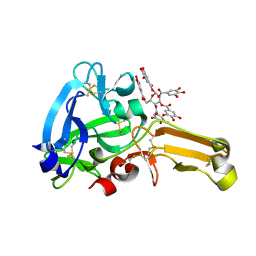 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai complex with tannic acid | | Descriptor: | 25 kDa polyphenol-binding protein, BETA-1,2,3,4,6-PENTA-O-GALLOYL-D-GLUCOPYRANOSE | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
5AXM
 
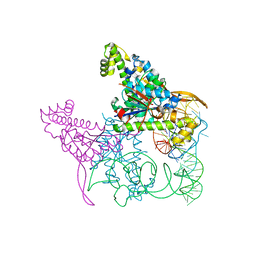 | | Crystal structure of Thg1 like protein (TLP) with tRNA(Phe) | | Descriptor: | MAGNESIUM ION, RNA (75-MER), tRNA(His)-5'-guanylyltransferase (Thg1) like protein | | Authors: | Kimura, S, Suzuki, T, Yu, J, Kato, K, Yao, M. | | Deposit date: | 2015-07-31 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Template-dependent nucleotide addition in the reverse (3'-5') direction by Thg1-like protein
Sci Adv, 2, 2016
|
|
5AXL
 
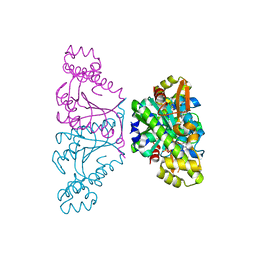 | | Crystal structure of Thg1 like protein (TLP) with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, tRNA(His)-5'-guanylyltransferase (Thg1) like protein | | Authors: | Kimura, S, Suzuki, T, Yu, J, Kato, K, Yao, M. | | Deposit date: | 2015-07-31 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Template-dependent nucleotide addition in the reverse (3'-5') direction by Thg1-like protein
Sci Adv, 2, 2016
|
|
5AXK
 
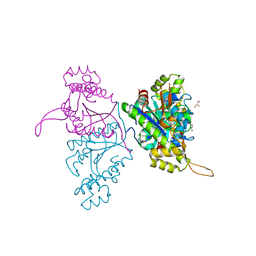 | | Crystal structure of Thg1 like protein (TLP) | | Descriptor: | GLYCEROL, tRNA(His)-5'-guanylyltransferase (Thg1) like protein | | Authors: | Kimura, S, Suzuki, T, Yu, J, Kato, K, Yao, M. | | Deposit date: | 2015-07-31 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Template-dependent nucleotide addition in the reverse (3'-5') direction by Thg1-like protein
Sci Adv, 2, 2016
|
|
1IR2
 
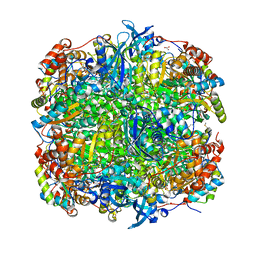 | | Crystal Structure of Activated Ribulose-1,5-bisphosphate Carboxylase/oxygenase (Rubisco) from Green alga, Chlamydomonas reinhardtii Complexed with 2-Carboxyarabinitol-1,5-bisphosphate (2-CABP) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, GLYCEROL, Large subunit of Rubisco, ... | | Authors: | Mizohata, E, Matsumura, H, Okano, Y, Kumei, M, Takuma, H, Onodera, J, Kato, K, Shibata, N, Inoue, T, Yokota, A, Kai, Y. | | Deposit date: | 2001-09-03 | | Release date: | 2002-03-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of activated ribulose-1,5-bisphosphate carboxylase/oxygenase from green alga Chlamydomonas reinhardtii complexed with 2-carboxyarabinitol-1,5-bisphosphate.
J.Mol.Biol., 316, 2002
|
|
6J3Z
 
 | | Structure of C2S1M1-type PSII-FCPII supercomplex from diatom | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nagao, R, Kato, K, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-01-07 | | Release date: | 2019-08-07 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for energy harvesting and dissipation in a diatom PSII-FCPII supercomplex.
Nat.Plants, 5, 2019
|
|
6J3Y
 
 | | Structure of C2S2-type PSII-FCPII supercomplex from diatom | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nagao, R, Kato, K, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-01-07 | | Release date: | 2019-08-07 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for energy harvesting and dissipation in a diatom PSII-FCPII supercomplex.
Nat.Plants, 5, 2019
|
|
6JPT
 
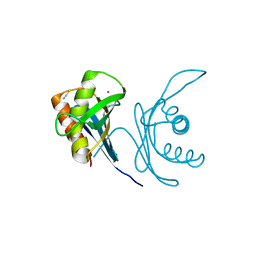 | | Crystal structure of human PAC3 homodimer (trigonal form) | | Descriptor: | POTASSIUM ION, Proteasome assembly chaperone 3, THIOCYANATE ION | | Authors: | Satoh, T, Yagi-Utsumi, M, Okamoto, K, Kurimoto, E, Tanaka, K, Kato, K. | | Deposit date: | 2019-03-27 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Molecular and Structural Basis of the Proteasome alpha Subunit Assembly Mechanism Mediated by the Proteasome-Assembling Chaperone PAC3-PAC4 Heterodimer.
Int J Mol Sci, 20, 2019
|
|
5H7L
 
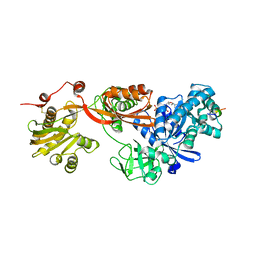 | | Complex of Elongation factor 2-50S ribosomal protein L12 | | Descriptor: | 50S ribosomal protein L12, Elongation factor 2, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Tanzawa, T, Kato, K, Uchiumi, T, Yao, M. | | Deposit date: | 2016-11-18 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The C-terminal helix of ribosomal P stalk recognizes a hydrophobic groove of elongation factor 2 in a novel fashion
Nucleic Acids Res., 46, 2018
|
|
7VD5
 
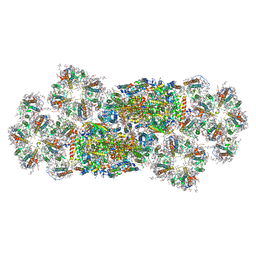 | | Structure of C2S2M2-type PSII-FCPII supercomplex from diatom | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Nagao, R, Kato, K, Akita, F, Miyazaki, N, Shen, J.R. | | Deposit date: | 2021-09-06 | | Release date: | 2022-03-02 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis for different types of hetero-tetrameric light-harvesting complexes in a diatom PSII-FCPII supercomplex
Nat Commun, 13, 2022
|
|
5H7J
 
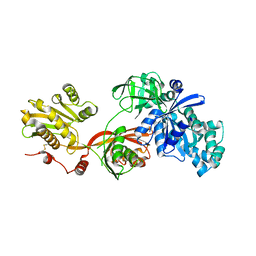 | | Crystal structure of Elongation factor 2 | | Descriptor: | Elongation factor 2, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Tanzawa, T, Kato, K, Uchiumi, T, Yao, M. | | Deposit date: | 2016-11-18 | | Release date: | 2018-02-21 | | Last modified: | 2018-05-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The C-terminal helix of ribosomal P stalk recognizes a hydrophobic groove of elongation factor 2 in a novel fashion
Nucleic Acids Res., 46, 2018
|
|
