1YEP
 
 | | Structural and biochemical analysis of the link between enzymatic activity and olgomerization in AhpC, a bacterial peroxiredoxin. | | Descriptor: | Alkyl hydroperoxide reductase subunit C, SULFATE ION | | Authors: | Parsonage, D, Youngblood, D.S, Sarma, G.N, Wood, Z.A, Karplus, P.A, Poole, L.B. | | Deposit date: | 2004-12-28 | | Release date: | 2005-08-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Analysis of the Link between Enzymatic Activity and Oligomeric State in AhpC, a Bacterial Peroxiredoxin.
Biochemistry, 44, 2005
|
|
3HVX
 
 | |
3I43
 
 | |
3HVS
 
 | |
3HVV
 
 | |
3GLW
 
 | |
1FNC
 
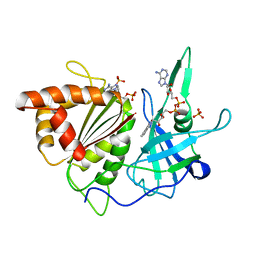 | | REFINED CRYSTAL STRUCTURE OF SPINACH FERREDOXIN REDUCTASE AT 1.7 ANGSTROMS RESOLUTION: OXIDIZED, REDUCED, AND 2'-PHOSPHO-5'-AMP BOUND STATES | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FERREDOXIN-NADP+ REDUCTASE, ... | | Authors: | Bruns, C.M, Karplus, P.A. | | Deposit date: | 1995-01-05 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined crystal structure of spinach ferredoxin reductase at 1.7 A resolution: oxidized, reduced and 2'-phospho-5'-AMP bound states.
J.Mol.Biol., 247, 1995
|
|
1FNB
 
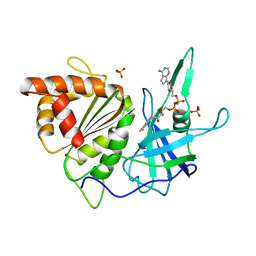 | | REFINED CRYSTAL STRUCTURE OF SPINACH FERREDOXIN REDUCTASE AT 1.7 ANGSTROMS RESOLUTION: OXIDIZED, REDUCED, AND 2'-PHOSPHO-5'-AMP BOUND STATES | | Descriptor: | FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Bruns, C.M, Karplus, P.A. | | Deposit date: | 1995-01-05 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined crystal structure of spinach ferredoxin reductase at 1.7 A resolution: oxidized, reduced and 2'-phospho-5'-AMP bound states.
J.Mol.Biol., 247, 1995
|
|
1FND
 
 | | REFINED CRYSTAL STRUCTURE OF SPINACH FERREDOXIN REDUCTASE AT 1.7 ANGSTROMS RESOLUTION: OXIDIZED, REDUCED, AND 2'-PHOSPHO-5'-AMP BOUND STATES | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bruns, C.M, Karplus, P.A. | | Deposit date: | 1995-01-05 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined crystal structure of spinach ferredoxin reductase at 1.7 A resolution: oxidized, reduced and 2'-phospho-5'-AMP bound states.
J.Mol.Biol., 247, 1995
|
|
1HYU
 
 | | CRYSTAL STRUCTURE OF INTACT AHPF | | Descriptor: | ALKYL HYDROPEROXIDE REDUCTASE SUBUNIT F, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Wood, Z.A, Poole, L.B, Karplus, P.A. | | Deposit date: | 2001-01-22 | | Release date: | 2001-02-14 | | Last modified: | 2012-05-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of intact AhpF reveals a mirrored thioredoxin-like active site and implies large domain rotations during catalysis.
Biochemistry, 40, 2001
|
|
1A5M
 
 | | K217A VARIANT OF KLEBSIELLA AEROGENES UREASE | | Descriptor: | UREASE (ALPHA SUBUNIT), UREASE (BETA SUBUNIT), UREASE (GAMMA SUBUNIT) | | Authors: | Pearson, M.A, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chemical rescue of Klebsiella aerogenes urease variants lacking the carbamylated-lysine nickel ligand.
Biochemistry, 37, 1998
|
|
1A5L
 
 | | K217C VARIANT OF KLEBSIELLA AEROGENES UREASE | | Descriptor: | UREASE (ALPHA SUBUNIT), UREASE (BETA SUBUNIT), UREASE (GAMMA SUBUNIT) | | Authors: | Pearson, M.A, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chemical rescue of Klebsiella aerogenes urease variants lacking the carbamylated-lysine nickel ligand.
Biochemistry, 37, 1998
|
|
1A5K
 
 | | K217E VARIANT OF KLEBSIELLA AEROGENES UREASE | | Descriptor: | UREASE (ALPHA SUBUNIT), UREASE (BETA SUBUNIT), UREASE (GAMMA SUBUNIT) | | Authors: | Pearson, M.A, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chemical rescue of Klebsiella aerogenes urease variants lacking the carbamylated-lysine nickel ligand.
Biochemistry, 37, 1998
|
|
1A5N
 
 | | K217A VARIANT OF KLEBSIELLA AEROGENES UREASE, CHEMICALLY RESCUED BY FORMATE AND NICKEL | | Descriptor: | FORMIC ACID, NICKEL (II) ION, UREASE (ALPHA SUBUNIT), ... | | Authors: | Pearson, M.A, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chemical rescue of Klebsiella aerogenes urease variants lacking the carbamylated-lysine nickel ligand.
Biochemistry, 37, 1998
|
|
1A5O
 
 | | K217C VARIANT OF KLEBSIELLA AEROGENES UREASE, CHEMICALLY RESCUED BY FORMATE AND NICKEL | | Descriptor: | FORMIC ACID, NICKEL (II) ION, UREASE (ALPHA SUBUNIT), ... | | Authors: | Pearson, M.A, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Chemical rescue of Klebsiella aerogenes urease variants lacking the carbamylated-lysine nickel ligand.
Biochemistry, 37, 1998
|
|
1K02
 
 | | Crystal Structure of Old Yellow Enzyme Mutant Gln114Asn | | Descriptor: | FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, NADPH DEHYDROGENASE 1 | | Authors: | Brown, B.J, Hyun, J, Duvvuri, S.D, Karplus, P.A, Massey, V. | | Deposit date: | 2001-09-18 | | Release date: | 2001-09-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The role of glutamine 114 in old yellow enzyme
J.Biol.Chem., 277, 2002
|
|
1K4Q
 
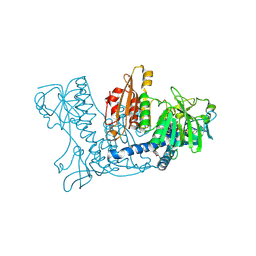 | | Human Glutathione Reductase Inactivated by Peroxynitrite | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glutathione Reductase | | Authors: | Savvides, S.N, Scheiwein, M, Boehme, C.C, Arteel, G.E, Karplus, P.A, Becker, K, Schirmer, R.H. | | Deposit date: | 2001-10-08 | | Release date: | 2002-01-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the antioxidant enzyme glutathione reductase inactivated by peroxynitrite.
J.Biol.Chem., 277, 2002
|
|
1K03
 
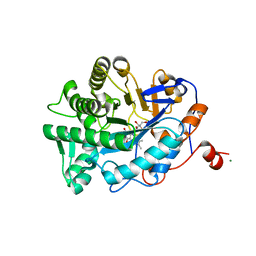 | | Crystal Structure of Old Yellow Enzyme Mutant Gln114Asn Complexed with Para-hydroxy Benzaldehyde | | Descriptor: | FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, NADPH DEHYDROGENASE 1, ... | | Authors: | Brown, B.J, Hyun, J, Duvvuri, S.D, Karplus, P.A, Massey, V. | | Deposit date: | 2001-09-18 | | Release date: | 2001-09-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The role of glutamine 114 in old yellow enzyme
J.Biol.Chem., 277, 2002
|
|
1JS4
 
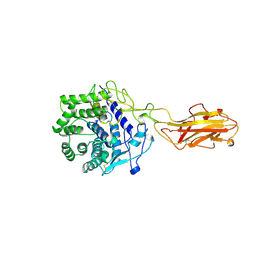 | | ENDO/EXOCELLULASE:CELLOBIOSE FROM THERMOMONOSPORA | | Descriptor: | CALCIUM ION, ENDO/EXOCELLULASE E4, beta-D-glucopyranose, ... | | Authors: | Sakon, J, Wilson, D.B, Karplus, P.A. | | Deposit date: | 1997-05-30 | | Release date: | 1997-09-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca.
Nat.Struct.Biol., 4, 1997
|
|
4PBR
 
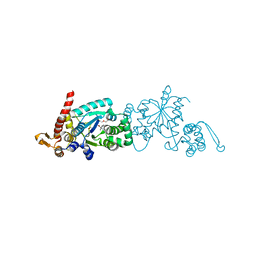 | | Crystal structure of the M. jannaschii G2 tRNA synthetase variant bound to 4-(2-bromoisobutyramido)-phenylalanine (BibaF) | | Descriptor: | 4-[(2-bromo-2-methylpropanoyl)amino]-L-phenylalanine, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Cooley, R.B, Karplus, P.A, Mehl, R.A. | | Deposit date: | 2014-04-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Gleaning Unexpected Fruits from Hard-Won Synthetases: Probing Principles of Permissivity in Non-canonical Amino Acid-tRNA Synthetases.
Chembiochem, 15, 2014
|
|
4PTZ
 
 | | Crystal structure of the Escherichia coli alkanesulfonate FMN reductase SsuE in FMN-bound form | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN reductase SsuE, GLYCEROL, ... | | Authors: | Driggers, C.M, Ellis, H.R, Karplus, P.A. | | Deposit date: | 2014-03-11 | | Release date: | 2014-06-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9007 Å) | | Cite: | Crystal Structure of Escherichia coli SsuE: Defining a General Catalytic Cycle for FMN Reductases of the Flavodoxin-like Superfamily.
Biochemistry, 53, 2014
|
|
4P53
 
 | | ValA (2-epi-5-epi-valiolone synthase) from Streptomyces hygroscopicus subsp. jinggangensis 5008 with NAD+ and Zn2+ bound | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Cyclase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kean, K.M, Codding, S.J, Karplus, P.A. | | Deposit date: | 2014-03-13 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a sedoheptulose 7-phosphate cyclase: ValA from Streptomyces hygroscopicus.
Biochemistry, 53, 2014
|
|
4QMA
 
 | | Crystal Structure of a Putative Cysteine Dioxygnase From Ralstonia eutropha: An Alternative Modeling of 2GM6 from JCSG Target 361076 | | Descriptor: | 1,2-ETHANEDIOL, Cysteine dioxygenase type I, FE (III) ION, ... | | Authors: | Hartman, S.H, Driggers, C.M, Karplus, P.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-06-15 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of Arg- and Gln-type bacterial cysteine dioxygenase homologs.
Protein Sci., 24, 2015
|
|
4PBT
 
 | | Crystal structure of the M. jannaschii G2 tRNA synthetase variant bound to 4-trans-cyclooctene-amidopheylalanine (Tco-amF) | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 4-{[(1R,4E)-cyclooct-4-en-1-ylcarbonyl]amino}-L-phenylalanine, GLYCEROL, ... | | Authors: | Cooley, R.B, Karplus, P.A, Mehl, R.A. | | Deposit date: | 2014-04-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Gleaning Unexpected Fruits from Hard-Won Synthetases: Probing Principles of Permissivity in Non-canonical Amino Acid-tRNA Synthetases.
Chembiochem, 15, 2014
|
|
4ND6
 
 | | Crystal structure of apo 3-nitro-tyrosine tRNA synthetase (5B) in the open form | | Descriptor: | GLYCEROL, Tyrosine-tRNA ligase | | Authors: | Cooley, R.B, Driggers, C.M, Karplus, P.A, Mehl, R.A. | | Deposit date: | 2013-10-25 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Improved Second-Generation 3-Nitro-tyrosine tRNA Synthetases.
Biochemistry, 53, 2014
|
|
