1B5U
 
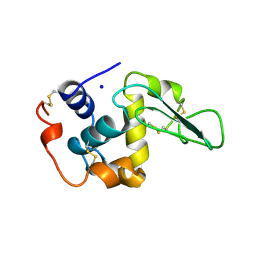 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANT | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
1B5Z
 
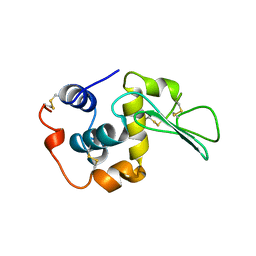 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANTS | | Descriptor: | LYSOZYME | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
1C43
 
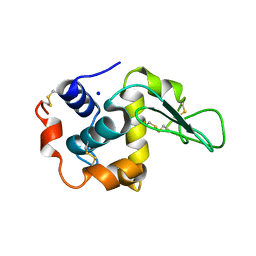 | | MUTANT HUMAN LYSOZYME WITH FOREIGN N-TERMINAL RESIDUES | | Descriptor: | PROTEIN (HUMAN LYSOZYME), SODIUM ION | | Authors: | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1999-08-03 | | Release date: | 1999-08-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effect of foreign N-terminal residues on the conformational stability of human lysozyme.
Eur.J.Biochem., 266, 1999
|
|
1C45
 
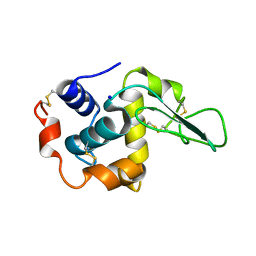 | | MUTANT HUMAN LYSOZYME WITH FOREIGN N-TERMINAL RESIDUES | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1999-08-03 | | Release date: | 1999-08-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effect of foreign N-terminal residues on the conformational stability of human lysozyme.
Eur.J.Biochem., 266, 1999
|
|
1B5W
 
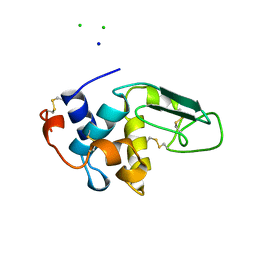 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANTS | | Descriptor: | CHLORIDE ION, PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
1C46
 
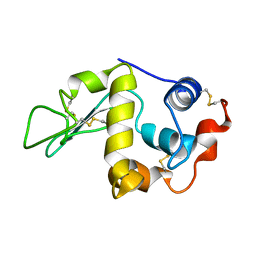 | |
1B5V
 
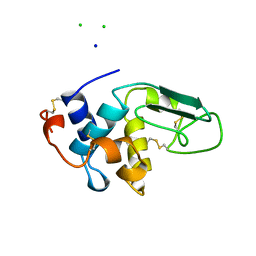 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANTS | | Descriptor: | CHLORIDE ION, PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-01-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
1B5Y
 
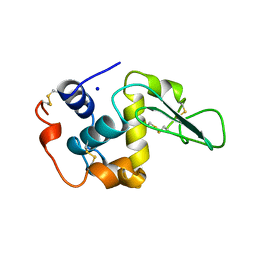 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANTS | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
5ZC0
 
 | |
1UCL
 
 | | Mutants of RNase Sa | | Descriptor: | Guanyl-specific ribonuclease Sa, SULFATE ION | | Authors: | Takano, K, Scholtz, J.M, Sacchettini, J.C, Pace, C.N. | | Deposit date: | 2003-04-15 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The contribution of polar group burial to protein stability is strongly context-dependent
J.Biol.Chem., 278, 2003
|
|
1UCI
 
 | | Mutants of RNase Sa | | Descriptor: | Guanyl-specific ribonuclease Sa, SULFATE ION | | Authors: | Takano, K, Scholtz, J.M, Sacchettini, J.C, Pace, C.N. | | Deposit date: | 2003-04-15 | | Release date: | 2003-09-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The contribution of polar group burial to protein stability is strongly context-dependent
J.Biol.Chem., 278, 2003
|
|
1UCK
 
 | | Mutants of RNase Sa | | Descriptor: | Guanyl-specific ribonuclease Sa, SULFATE ION | | Authors: | Takano, K, Scholtz, J.M, Sacchettini, J.C, Pace, C.N. | | Deposit date: | 2003-04-15 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The contribution of polar group burial to protein stability is strongly context-dependent
J.Biol.Chem., 278, 2003
|
|
1CKC
 
 | | T43A MUTANT HUMAN LYSOZYME | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Funahashi, J, Yutani, K. | | Deposit date: | 1999-04-22 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of intra- and intermolecular hydrogen bonds to the conformational stability of human lysozyme(,).
Biochemistry, 38, 1999
|
|
1UCJ
 
 | | Mutants of RNase Sa | | Descriptor: | Guanyl-specific ribonuclease Sa, SULFATE ION | | Authors: | Takano, K, Scholtz, J.M, Sacchettini, J.C, Pace, C.N. | | Deposit date: | 2003-04-15 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The contribution of polar group burial to protein stability is strongly context-dependent
J.Biol.Chem., 278, 2003
|
|
1IP7
 
 | | G129A HUMAN LYSOZYME | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2001-04-20 | | Release date: | 2001-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
1IP5
 
 | | G105A HUMAN LYSOZYME | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2001-04-20 | | Release date: | 2001-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
1IP4
 
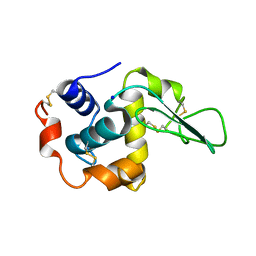 | | G72A HUMAN LYSOZYME | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2001-04-20 | | Release date: | 2001-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
1IP3
 
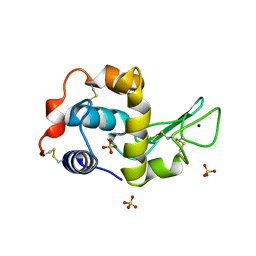 | | G68A HUMAN LYSOZYME | | Descriptor: | LYSOZYME C, SODIUM ION, SULFATE ION | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2001-04-20 | | Release date: | 2001-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
1IP2
 
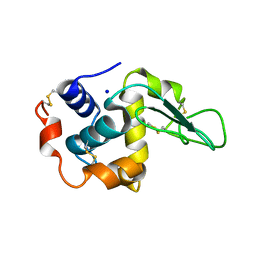 | | G48A HUMAN LYSOZYME | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2001-04-20 | | Release date: | 2001-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
1IX0
 
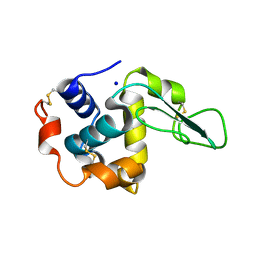 | | I59A-3SS human lysozyme | | Descriptor: | SODIUM ION, lysozyme | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2002-06-06 | | Release date: | 2003-07-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Buried water molecules contribute to the conformational stability of a protein
PROTEIN ENG., 16, 2003
|
|
1IP6
 
 | | G127A HUMAN LYSOZYME | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2001-04-20 | | Release date: | 2001-11-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
1IP1
 
 | | G37A HUMAN LYSOZYME | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2001-04-20 | | Release date: | 2001-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
7D5M
 
 | | Crystal structure of inositol dehydrogenase homolog complexed with NAD+ from Azotobacter vinelandii | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Oxidoreductase | | Authors: | Fukano, K, Ono, T, Suzuki, M, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-09-27 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of inositol dehydrogenase complexed with NAD+ from Azotobacter vinelandii
To Be Published
|
|
7D5N
 
 | | Crystal structure of inositol dehydrogenase homolog complexed with NADH and myo-inositol from Azotobacter vinelandii | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Oxidoreductase | | Authors: | Fukano, K, Ono, T, Suzuki, M, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-09-27 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of inositol dehydrogenase complexed with NADH and myo-inositol from Azotobacter vinelandii
To Be Published
|
|
1X1P
 
 | | Crystal structure of Tk-RNase HII(1-197)-A(28-42) | | Descriptor: | Ribonuclease HII | | Authors: | Takano, K, Endo, S, Mukaiyama, A, Chon, H, Matsumura, H, Koga, Y, Kanaya, S. | | Deposit date: | 2005-04-11 | | Release date: | 2006-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of amyloid beta fragments in aqueous environments
Febs J., 273, 2006
|
|
