4WPZ
 
 | | Crystal structure of cytochrome P450 CYP107W1 from Streptomyces avermitilis | | Descriptor: | Cytochrome P450, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kang, L.W, Kim, D.H, Pham, T.V, Han, S.H. | | Deposit date: | 2014-10-21 | | Release date: | 2015-04-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional characterization of CYP107W1 from Streptomyces avermitilis and biosynthesis of macrolide oligomycin A.
Arch.Biochem.Biophys., 575, 2015
|
|
4WQ0
 
 | |
3HE8
 
 | | Structural study of Clostridium thermocellum Ribose-5-Phosphate Isomerase B | | Descriptor: | GLYCEROL, Ribose-5-phosphate isomerase | | Authors: | Kang, L.W, Kim, J.K, Jung, J.H, Hong, M.K. | | Deposit date: | 2009-05-08 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Clostridium thermocellum ribose-5-phosphate isomerase B reveals properties critical for fast enzyme kinetics.
Appl.Microbiol.Biotechnol., 90, 2011
|
|
3HEE
 
 | | Structural study of Clostridium thermocellum Ribose-5-Phosphate Isomerase B and ribose-5-phosphate | | Descriptor: | RIBOSE-5-PHOSPHATE, Ribose-5-phosphate isomerase | | Authors: | Kang, L.W, Kim, J.K, Jung, J.H, Hong, M.K. | | Deposit date: | 2009-05-08 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Clostridium thermocellum ribose-5-phosphate isomerase B reveals properties critical for fast enzyme kinetics.
Appl.Microbiol.Biotechnol., 90, 2011
|
|
2O4N
 
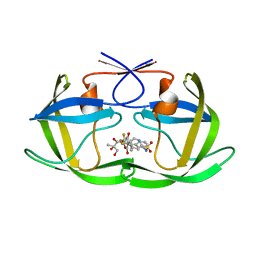 | | Crystal Structure of HIV-1 Protease (TRM Mutant) in Complex with Tipranavir | | Descriptor: | GLYCEROL, N-(3-{(1R)-1-[(6R)-4-HYDROXY-2-OXO-6-PHENETHYL-6-PROPYL-5,6-DIHYDRO-2H-PYRAN-3-YL]PROPYL}PHENYL)-5-(TRIFLUOROMETHYL)-2-PYRIDINESULFONAMIDE, protease | | Authors: | Kang, L.W, Armstrong, A.A, Muzammil, S, Jakalian, A, Bonneau, P.R, Schmelmer, V, Freire, E, Amzel, L.M. | | Deposit date: | 2006-12-04 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unique thermodynamic response of tipranavir to human immunodeficiency virus type 1 protease drug resistance mutations.
J.Virol., 81, 2007
|
|
2O4P
 
 | | Crystal Structure of HIV-1 Protease (Q7K) in Complex with Tipranavir | | Descriptor: | GLYCEROL, N-(3-{(1R)-1-[(6R)-4-HYDROXY-2-OXO-6-PHENETHYL-6-PROPYL-5,6-DIHYDRO-2H-PYRAN-3-YL]PROPYL}PHENYL)-5-(TRIFLUOROMETHYL)-2-PYRIDINESULFONAMIDE, protease | | Authors: | Kang, L.W, Armstrong, A.A, Muzammil, S, Jakalian, A, Bonneau, P.R, Schmelmer, V, Freire, E, Amzel, L.M. | | Deposit date: | 2006-12-04 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unique thermodynamic response of tipranavir to human immunodeficiency virus type 1 protease drug resistance mutations.
J.Virol., 81, 2007
|
|
1NQY
 
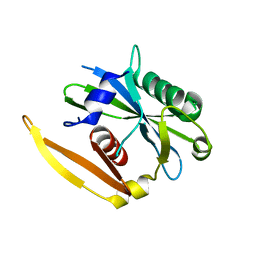 | | The structure of a CoA pyrophosphatase from D. Radiodurans | | Descriptor: | CoA pyrophosphatase (MutT/nudix family protein) | | Authors: | Kang, L.W, Gabelli, S.B, Bianchet, M.A, Xu, W.L, Bessman, M.J, Amzel, L.M. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of a coenzyme A pyrophosphatase from Deinococcus radiodurans: a member of the Nudix family.
J.Bacteriol., 185, 2003
|
|
1NQZ
 
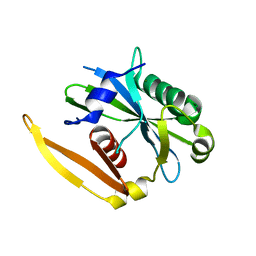 | | The structure of a CoA pyrophosphatase from D. Radiodurans complexed with a magnesium ion | | Descriptor: | CoA pyrophosphatase (MutT/nudix family protein), MAGNESIUM ION | | Authors: | Kang, L.W, Gabelli, S.B, Bianchet, M.A, Xu, W.L, Bessman, M.J, Amzel, L.M. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a coenzyme A pyrophosphatase from Deinococcus radiodurans: a member of the Nudix family.
J.Bacteriol., 185, 2003
|
|
3E6G
 
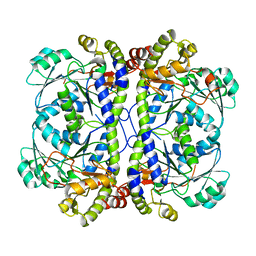 | | Crystal structure of XometC, a cystathionine c-lyase-like protein from Xanthomonas oryzae pv.oryzae | | Descriptor: | Cystathionine gamma-lyase-like protein | | Authors: | Ngo, H.P.T, Kim, J.K, Kim, H.S, Jung, J.H, Ahn, Y.J, Kim, J.G, Lee, B.M, Kang, H.W, Kang, L.W. | | Deposit date: | 2008-08-15 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of XometC, a cystathionine c-lyase-like protein from Xanthomonas oryzae pv.oryzae
To be published
|
|
3K89
 
 | |
4ME6
 
 | | Crystal structure of D-alanine-D-alanine ligase A from Xanthomonas oryzae pathovar oryzae with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-alanine--D-alanine ligase, MAGNESIUM ION | | Authors: | Doan, T.T.N, Kim, J.K, Ahn, Y.J, Lee, B.M, Kang, L.W. | | Deposit date: | 2013-08-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of d-alanine-d-alanine ligase from Xanthomonas oryzae pv. oryzae alone and in complex with nucleotides
Arch.Biochem.Biophys., 545C, 2014
|
|
8YQ1
 
 | |
6LD9
 
 | |
6LD7
 
 | |
9IXO
 
 | | Crystal structure of OXA-14 | | Descriptor: | Beta-lactamase | | Authors: | Lee, C.E, Park, Y.S, Park, H.J, Kang, L.W. | | Deposit date: | 2024-07-29 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural insights into alterations in the substrate spectrum of serine-beta-lactamase OXA-10 from Pseudomonas aeruginosa by single amino acid substitutions.
Emerg Microbes Infect, 13, 2024
|
|
9IXP
 
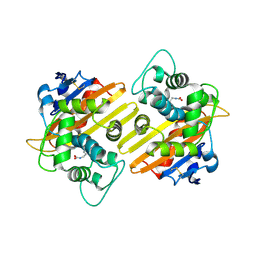 | | Crystal structure of OXA-10 variant A124T | | Descriptor: | Beta-lactamase OXA-10 | | Authors: | Lee, C.E, Park, Y.S, Park, H.J, Kang, L.W. | | Deposit date: | 2024-07-29 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into alterations in the substrate spectrum of serine-beta-lactamase OXA-10 from Pseudomonas aeruginosa by single amino acid substitutions.
Emerg Microbes Infect, 13, 2024
|
|
9IXR
 
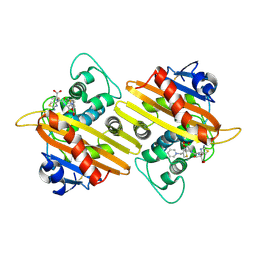 | | Crystal structure of OXA-10 variant A124T in the complex with ceftazidime | | Descriptor: | 1-({(2R)-2-[(1R)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-2-oxoethyl]-4-carboxy-3,6-dihydro-2H-1,3-thiazin-5-yl}methyl)pyridinium, Beta-lactamase OXA-10 | | Authors: | Lee, C.E, Park, Y.S, Park, H.J, Kang, L.W. | | Deposit date: | 2024-07-29 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural insights into alterations in the substrate spectrum of serine-beta-lactamase OXA-10 from Pseudomonas aeruginosa by single amino acid substitutions.
Emerg Microbes Infect, 13, 2024
|
|
9IXN
 
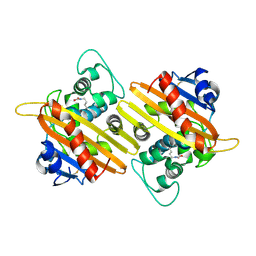 | | Crystal structure of OXA-10 | | Descriptor: | Beta-lactamase OXA-10 | | Authors: | Lee, C.E, Park, Y.S, Park, H.J, Kang, L.W. | | Deposit date: | 2024-07-29 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural insights into alterations in the substrate spectrum of serine-beta-lactamase OXA-10 from Pseudomonas aeruginosa by single amino acid substitutions.
Emerg Microbes Infect, 13, 2024
|
|
9IXQ
 
 | | Crystal structure of OXA-17 | | Descriptor: | Beta-lactamase | | Authors: | Lee, C.E, Park, Y.S, Park, H.J, Kang, L.W. | | Deposit date: | 2024-07-29 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural insights into alterations in the substrate spectrum of serine-beta-lactamase OXA-10 from Pseudomonas aeruginosa by single amino acid substitutions.
Emerg Microbes Infect, 13, 2024
|
|
3DKU
 
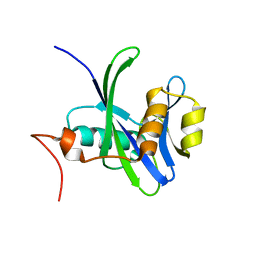 | | Crystal structure of Nudix hydrolase Orf153, ymfB, from Escherichia coli K-1 | | Descriptor: | Putative phosphohydrolase | | Authors: | Hong, M.K, Kim, J.K, Jung, J.H, Jung, J.W, Choi, J.Y, Kang, L.W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of Nudix hydrolase Orf153, ymfB, from Escherichia coli K-1.
To be Published
|
|
3E5N
 
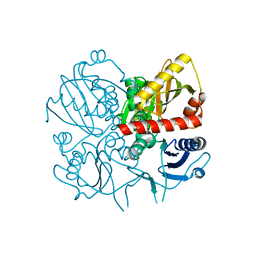 | | Crystal structure of D-alanine-D-alanine ligase from Xanthomonas oryzae pv. oryzae KACC10331 | | Descriptor: | D-alanine-D-alanine ligase A | | Authors: | Doan, T.N.T, Kim, J.K, Kim, H.S, Ahn, Y.J, Kim, J.G, Lee, B.M, Kang, L.W. | | Deposit date: | 2008-08-14 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of D-alanine-D-alanine ligase from Xanthomonas oryzae pv. oryzae KACC10331
To be published
|
|
3UU0
 
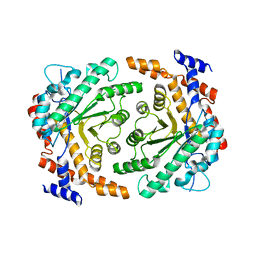 | | Crystal structure of L-rhamnose isomerase from Bacillus halodurans in complex with Mn | | Descriptor: | L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Doan, T.T.N, Prabhu, P, Kim, J.K, Jeya, M, Kang, L.W, Lee, J.K. | | Deposit date: | 2011-11-27 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based studies on the metal binding of two-metal-dependent sugar isomerases.
Febs J., 281, 2014
|
|
3UVA
 
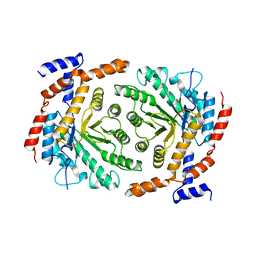 | | Crystal structure of L-rhamnose isomerase mutant W38F from Bacillus halodurans in complex with Mn | | Descriptor: | L-Rhamnose isomerase, MANGANESE (II) ION | | Authors: | Doan, T.T.N, Prabhu, P, Jeya, M, Kim, J.K, Kang, L.W, Lee, J.K. | | Deposit date: | 2011-11-29 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure-based studies on the metal binding of two-metal-dependent sugar isomerases.
Febs J., 281, 2014
|
|
3UXI
 
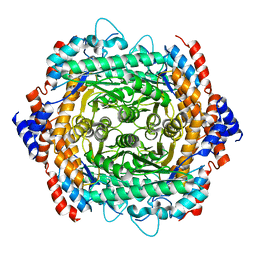 | | Crystal structure of L-rhamnose isomerase W38A mutant from Bacillus halodurans | | Descriptor: | L-Rhamnose isomerase, MANGANESE (II) ION | | Authors: | Doan, T.T.N, Prabhu, P, Kim, J.K, Jeya, M, Kang, L.W, Lee, J.K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-12-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure-based studies on the metal binding of two-metal-dependent sugar isomerases.
Febs J., 281, 2014
|
|
3FK5
 
 | |
