2N7R
 
 | |
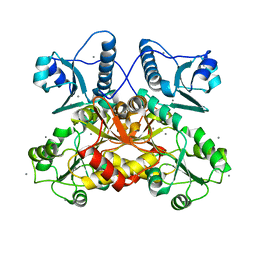
5CRH
 
 | |
5DQP
 
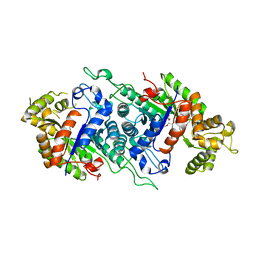 | | EDTA monooxygenase (EmoA) from Chelativorans sp. BNC1 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, EDTA monooxygenase, SULFATE ION | | Authors: | Jun, S.Y, Youn, B, Xun, L, Kang, C, Lewis, K.M. | | Deposit date: | 2015-09-15 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.146 Å) | | Cite: | Structural and biochemical characterization of EDTA monooxygenase and its physical interaction with a partner flavin reductase.
Mol.Microbiol., 100, 2016
|
|
5CRD
 
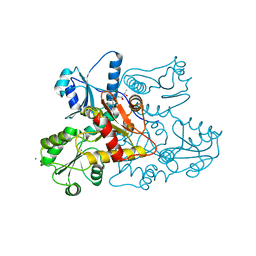 | | Wild-type human skeletal calsequestrin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Calsequestrin-1 | | Authors: | Lewis, K.M, Ronish, L.A, Kang, C. | | Deposit date: | 2015-07-22 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Characterization of Two Human Skeletal Calsequestrin Mutants Implicated in Malignant Hyperthermia and Vacuolar Aggregate Myopathy.
J.Biol.Chem., 290, 2015
|
|
5CRE
 
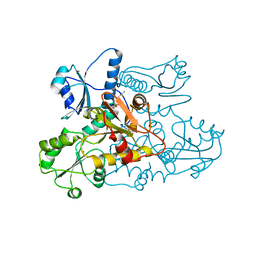 | | Human skeletal calsequestrin, D210G mutant low-calcium complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Calsequestrin-1 | | Authors: | Lewis, K.M, Ronish, L.A, Kang, C. | | Deposit date: | 2015-07-22 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.315 Å) | | Cite: | Characterization of Two Human Skeletal Calsequestrin Mutants Implicated in Malignant Hyperthermia and Vacuolar Aggregate Myopathy.
J.Biol.Chem., 290, 2015
|
|
5CRG
 
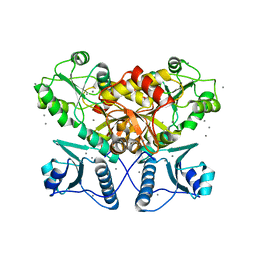 | |
8FF7
 
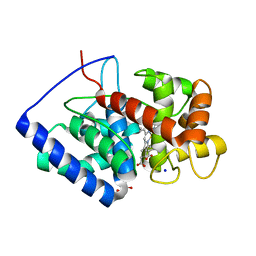 | |
8FF6
 
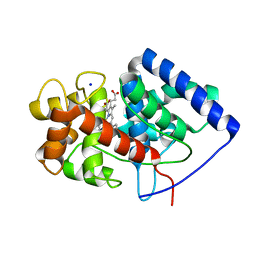 | | Cytosolic ascorbate peroxidase mutant from Panicum virgatum | | Descriptor: | Cytosolic ascorbate peroxidase, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-12-07 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Activity of Cytosolic Ascorbate Peroxidase (APX) from Panicum virgatum against Ascorbate and Phenylpropanoids.
Int J Mol Sci, 24, 2023
|
|
8FEM
 
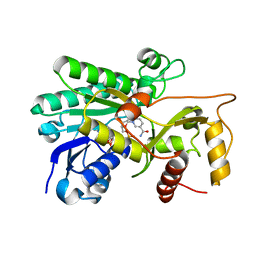 | | Panicum vigratum Dihydroflavonol 4-reductase complexed with NADP | | Descriptor: | Dihydroflavonol 4-Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lewis, J.A, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8FET
 
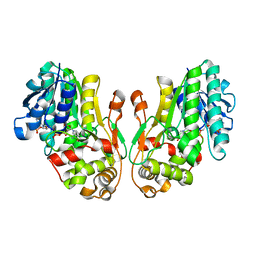 | | Flavanone 4-Reductase from Sorghum bicolor-NADP(H) complex | | Descriptor: | 3-deoxyanthocyanidin synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8FEV
 
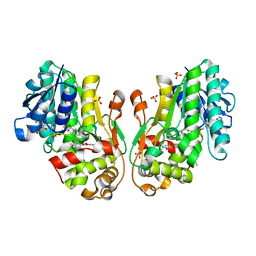 | | Flavanone 4-Reductase from Sorghum bicolor-NADP(H) and dihydroquercetin complex | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, 3-deoxyanthocyanidin synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8FIO
 
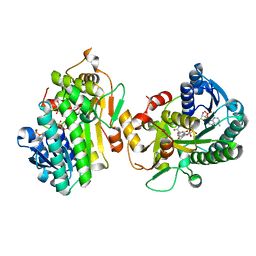 | | Hypothetical anthocyanidin reductase from Sorghum bicolor-NADP(H) and naringenin complex | | Descriptor: | Epimerase domain-containing protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NARINGENIN | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-12-16 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8FEW
 
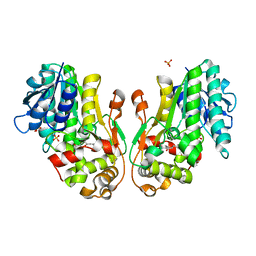 | | Flavanone 4-Reductase from Sorghum bicolor-naringenin complex | | Descriptor: | 3-deoxyanthocyanidin synthase, NARINGENIN, SULFATE ION | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8FEN
 
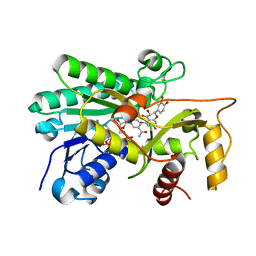 | | Panicum vigratum Dihydroflavonol 4-reductase complexed with NADP and DHQ | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Panicum virgatum Dihydroflavonol 4-Reductase | | Authors: | Lewis, J.A, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8FIP
 
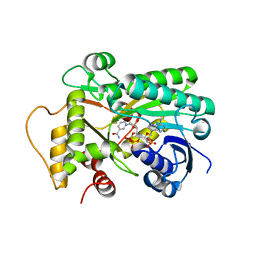 | | Hypothetical anthocyanidin reducatase from Sorghum bicolor- NADP+ complex | | Descriptor: | Epimerase domain-containing protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-12-16 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8FEU
 
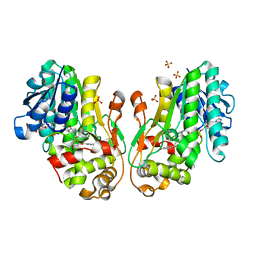 | | Flavanone 4-Reductase from Sorghum bicolor-NADP(H) and naringenin complex | | Descriptor: | 3-deoxyanthocyanidin synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NARINGENIN, ... | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
1CN0
 
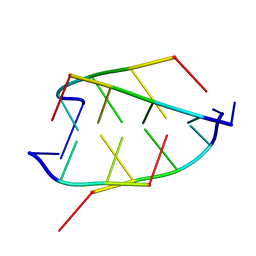 | | CRYSTAL STRUCTURE OF D(ACCCT) | | Descriptor: | DNA (5'-D(*AP*CP*CP*CP*T)-3') | | Authors: | Weil, J, Min, T, Cheng, Y, Wang, S, Sutherland, C, Sinha, N, Kang, C. | | Deposit date: | 1999-05-24 | | Release date: | 2000-05-24 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Stabilization of the i-motif by intramolecular adenine-adenine-thymine base triple in the structure of d(ACCCT).
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1FHM
 
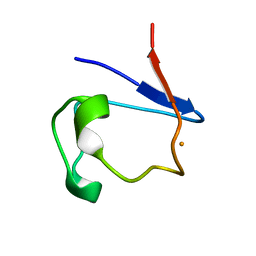 | | X-RAY CRYSTAL STRUCTURE OF REDUCED RUBREDOXIN | | Descriptor: | FE (II) ION, RUBREDOXIN | | Authors: | Min, T, Ergenekan, C.E, Eidsness, M.K, Ichiye, T, Kang, C. | | Deposit date: | 2000-08-02 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Leucine 41 is a gate for water entry in the reduction of Clostridium pasteurianum rubredoxin.
Protein Sci., 10, 2001
|
|
1FHH
 
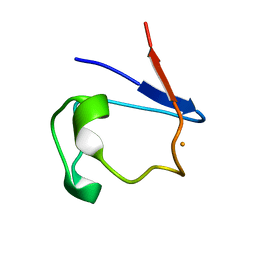 | | X-RAY CRYSTAL STRUCTURE OF OXIDIZED RUBREDOXIN | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Min, T, Ergenekan, C.E, Eidsness, M.K, Ichiye, T, Kang, C. | | Deposit date: | 2000-08-01 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Leucine 41 is a gate for water entry in the reduction of Clostridium pasteurianum rubredoxin.
Protein Sci., 10, 2001
|
|
4YSB
 
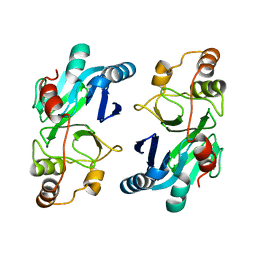 | | Crystal structure of ETHE1 from Myxococcus xanthus | | Descriptor: | FE (III) ION, Metallo-beta-lactamase family protein | | Authors: | Sattler, S.A, Wang, X, DeHan, P.J, Xun, L, Kang, C. | | Deposit date: | 2015-03-17 | | Release date: | 2015-06-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5015 Å) | | Cite: | Characterizations of Two Bacterial Persulfide Dioxygenases of the Metallo-beta-lactamase Superfamily.
J.Biol.Chem., 290, 2015
|
|
4YSK
 
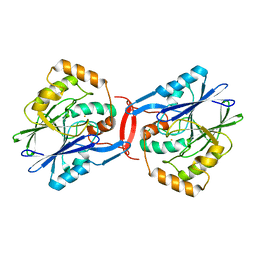 | | Crystal structure of apo-form SdoA from Pseudomonas putida | | Descriptor: | Beta-lactamase domain protein, FE (III) ION | | Authors: | Sattler, S.A, Wang, X, DeHan, P.J, Xun, L, Kang, C. | | Deposit date: | 2015-03-17 | | Release date: | 2015-06-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.466 Å) | | Cite: | Characterizations of Two Bacterial Persulfide Dioxygenases of the Metallo-beta-lactamase Superfamily.
J.Biol.Chem., 290, 2015
|
|
4YSL
 
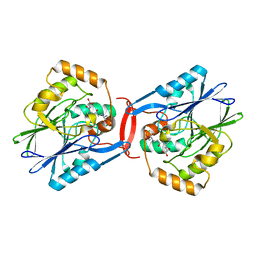 | | Crystal structure of SdoA from Pseudomonas putida in complex with glutathione | | Descriptor: | Beta-lactamase domain protein, FE (III) ION, GLUTATHIONE | | Authors: | Sattler, S.A, Wang, X, DeHan, P.J, Xun, L, Kang, C. | | Deposit date: | 2015-03-17 | | Release date: | 2015-06-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4618 Å) | | Cite: | Characterizations of Two Bacterial Persulfide Dioxygenases of the Metallo-beta-lactamase Superfamily.
J.Biol.Chem., 290, 2015
|
|
5ZMA
 
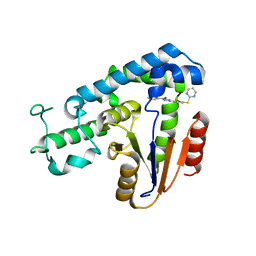 | | Structural basis for an allosteric Eya2 phosphatase inhibitor | | Descriptor: | 3-fluoro-N'-[(E)-{5-[(pyrimidin-2-yl)sulfanyl]furan-2-yl}methylidene]benzohydrazide, Eyes absent homolog 2 | | Authors: | Anantharajan, J, Jansson, A.E, Kang, C. | | Deposit date: | 2018-04-02 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.175 Å) | | Cite: | Structural and Functional Analyses of an Allosteric EYA2 Phosphatase Inhibitor That Has On-Target Effects in Human Lung Cancer Cells.
Mol.Cancer Ther., 18, 2019
|
|
6AFK
 
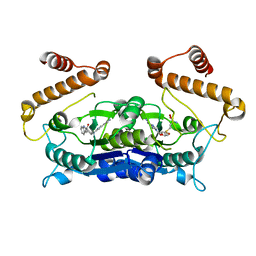 | | Crystal structure of TrmD from Pseudomonas aeruginosa in complex with active-site inhibitor | | Descriptor: | N-{(3S)-1-[3-(pyridin-4-yl)-1H-pyrazol-5-yl]piperidin-3-yl}-1H-indole-2-carboxamide, S-ADENOSYLMETHIONINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Zhong, W, Koay, A, Wong, Y.W, Sahili, A.E, Nah, Q, Kang, C, Poulsen, A, Chionh, Y.K, McBee, M, Matter, A, Hill, J, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Targeting the Bacterial Epitranscriptome for Antibiotic Development: Discovery of Novel tRNA-(N1G37) Methyltransferase (TrmD) Inhibitors.
Acs Infect Dis., 5, 2019
|
|
6AT7
 
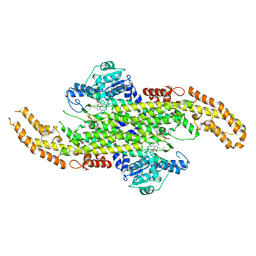 | | Phenylalanine Ammonia-Lyase (PAL) from Sorghum bicolor | | Descriptor: | AMMONIUM ION, Phenylalanine ammonia-lyase | | Authors: | Jun, S.Y, Kang, C. | | Deposit date: | 2017-08-28 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Biochemical and Structural Analysis of Substrate Specificity of a Phenylalanine Ammonia-Lyase.
Plant Physiol., 176, 2018
|
|
