6L3A
 
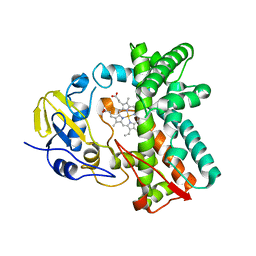 | | Cytochrome P450 107G1 (RapN) with everolimus | | Descriptor: | Cytochrome P450, Everolimus, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Km, V.C, Kim, D.H, Lim, Y.R, Lee, I.H, Lee, J.H, Kang, L.W. | | Deposit date: | 2019-10-10 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into CYP107G1 from rapamycin-producing Streptomyces rapamycinicus.
Arch.Biochem.Biophys., 692, 2020
|
|
6ILA
 
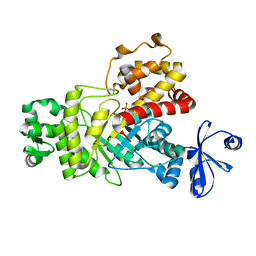 | | Two Glycerol complexed Crystal structure of fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi | | Descriptor: | Fructuronate-tagaturonate epimerase UxaE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Choi, M.Y, Kang, L.W, Ho, T.H, Nguyen, D.Q, Lee, I.H, Lee, J.H, Park, Y.S, Park, H.J. | | Deposit date: | 2018-10-17 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi
To be published
|
|
6JF3
 
 | |
6JFC
 
 | |
6L39
 
 | | Cytochrome P450 107G1 (RapN) | | Descriptor: | Cytochrome P450, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Kim, V.C, Kim, D.H, Lim, Y.R, Lee, I.H, Lee, J.H, Kang, L.W. | | Deposit date: | 2019-10-10 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural insights into CYP107G1 from rapamycin-producing Streptomyces rapamycinicus.
Arch.Biochem.Biophys., 692, 2020
|
|
9IXN
 
 | | Crystal structure of OXA-10 | | Descriptor: | Beta-lactamase OXA-10 | | Authors: | Lee, C.E, Park, Y.S, Park, H.J, Kang, L.W. | | Deposit date: | 2024-07-29 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Insights into Alterations in the Substrate Spectrum of Serine-beta-Lactamase OXA-10 from Pseudomonas aeruginosa by Single Amino Acid Substitutions.
Emerg Microbes Infect, 2024
|
|
9IXO
 
 | | Crystal structure of OXA-14 | | Descriptor: | Beta-lactamase | | Authors: | Lee, C.E, Park, Y.S, Park, H.J, Kang, L.W. | | Deposit date: | 2024-07-29 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Insights into Alterations in the Substrate Spectrum of Serine-beta-Lactamase OXA-10 from Pseudomonas aeruginosa by Single Amino Acid Substitutions.
Emerg Microbes Infect, 2024
|
|
9IXP
 
 | | Crystal structure of OXA-10 variant A124T | | Descriptor: | Beta-lactamase OXA-10 | | Authors: | Lee, C.E, Park, Y.S, Park, H.J, Kang, L.W. | | Deposit date: | 2024-07-29 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into Alterations in the Substrate Spectrum of Serine-beta-Lactamase OXA-10 from Pseudomonas aeruginosa by Single Amino Acid Substitutions.
Emerg Microbes Infect, 2024
|
|
9IXQ
 
 | | Crystal structure of OXA-17 | | Descriptor: | Beta-lactamase | | Authors: | Lee, C.E, Park, Y.S, Park, H.J, Kang, L.W. | | Deposit date: | 2024-07-29 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Insights into Alterations in the Substrate Spectrum of Serine-beta-Lactamase OXA-10 from Pseudomonas aeruginosa by Single Amino Acid Substitutions.
Emerg Microbes Infect, 2024
|
|
9IXR
 
 | | Crystal structure of OXA-10 variant A124T in the complex with ceftazidime | | Descriptor: | 1-({(2R)-2-[(1R)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-2-oxoethyl]-4-carboxy-3,6-dihydro-2H-1,3-thiazin-5-yl}methyl)pyridinium, Beta-lactamase OXA-10 | | Authors: | Lee, C.E, Park, Y.S, Park, H.J, Kang, L.W. | | Deposit date: | 2024-07-29 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Insights into Alterations in the Substrate Spectrum of Serine-beta-Lactamase OXA-10 from Pseudomonas aeruginosa by Single Amino Acid Substitutions.
Emerg Microbes Infect, 2024
|
|
4FXB
 
 | | Crystal structure of CYP105N1 from Streptomyces coelicolor: a cytochrome P450 oxidase in the coelibactin siderophore biosynthetic pathway | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 | | Authors: | Hong, M.K, Lim, Y.R, Kim, J.K, Kim, D.H, Kang, L.W. | | Deposit date: | 2012-07-03 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of cytochrome P450 CYP105N1 from Streptomyces coelicolor, an oxidase in the coelibactin siderophore biosynthetic pathway
Arch.Biochem.Biophys., 528, 2012
|
|
5I8U
 
 | | Crystal Structure of the RV1700 (MT ADPRASE) E142Q mutant | | Descriptor: | ADP-ribose pyrophosphatase, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Thirawatananond, P, Kang, L.-W, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2016-02-19 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and mutational studies of the adenosine diphosphate ribose hydrolase from Mycobacterium tuberculosis.
J. Bioenerg. Biomembr., 48, 2016
|
|
3DKU
 
 | | Crystal structure of Nudix hydrolase Orf153, ymfB, from Escherichia coli K-1 | | Descriptor: | Putative phosphohydrolase | | Authors: | Hong, M.K, Kim, J.K, Jung, J.H, Jung, J.W, Choi, J.Y, Kang, L.W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of Nudix hydrolase Orf153, ymfB, from Escherichia coli K-1.
To be Published
|
|
3P14
 
 | |
3SHD
 
 | |
5CP0
 
 | |
3E5N
 
 | | Crystal structure of D-alanine-D-alanine ligase from Xanthomonas oryzae pv. oryzae KACC10331 | | Descriptor: | D-alanine-D-alanine ligase A | | Authors: | Doan, T.N.T, Kim, J.K, Kim, H.S, Ahn, Y.J, Kim, J.G, Lee, B.M, Kang, L.W. | | Deposit date: | 2008-08-14 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of D-alanine-D-alanine ligase from Xanthomonas oryzae pv. oryzae KACC10331
To be published
|
|
4IY7
 
 | | crystal structure of cystathionine gamma lyase (XometC) from Xanthomonas oryzae pv. oryzae in complex with E-site serine, A-site external aldimine structure with serine and A-site external aldimine structure with aminoacrylate intermediates | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, Cystathionine gamma-lyase-like protein, ... | | Authors: | Ngo, H.P.T, Kim, J.K, Kang, L.W. | | Deposit date: | 2013-01-28 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | PLP undergoes conformational changes during the course of an enzymatic reaction.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NFW
 
 | | Structure and atypical hydrolysis mechanism of the Nudix hydrolase Orf153 (YmfB) from Escherichia coli | | Descriptor: | MANGANESE (II) ION, Putative Nudix hydrolase ymfB, SULFATE ION | | Authors: | Hong, M.K, Kim, J.K, Kang, L.W. | | Deposit date: | 2013-11-01 | | Release date: | 2014-05-14 | | Last modified: | 2015-03-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Divalent metal ion-based catalytic mechanism of the Nudix hydrolase Orf153 (YmfB) from Escherichia coli
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7BZ4
 
 | |
7BYQ
 
 | |
7BZ1
 
 | |
7BZI
 
 | |
7BZ3
 
 | |
3FK5
 
 | |
