4ME6
 
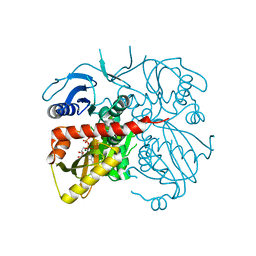 | | Crystal structure of D-alanine-D-alanine ligase A from Xanthomonas oryzae pathovar oryzae with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-alanine--D-alanine ligase, MAGNESIUM ION | | Authors: | Doan, T.T.N, Kim, J.K, Ahn, Y.J, Lee, B.M, Kang, L.W. | | Deposit date: | 2013-08-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of d-alanine-d-alanine ligase from Xanthomonas oryzae pv. oryzae alone and in complex with nucleotides
Arch.Biochem.Biophys., 545C, 2014
|
|
4NT8
 
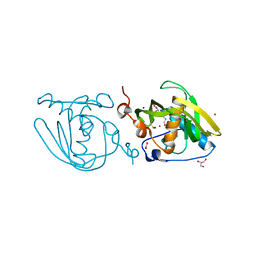 | | Formyl-methionine-alanine complex structure of peptide deformylase from Xanthomoonas oryzae pv. oryzae | | Descriptor: | ACETATE ION, ALANINE, CADMIUM ION, ... | | Authors: | Ngo, H.P.T, Kim, J.K, Kang, L.W. | | Deposit date: | 2013-12-02 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate complex structure of Xoo1075, a peptide deformylase, from Xanthomonas oryzae pv. oryzae
To be Published
|
|
4QD4
 
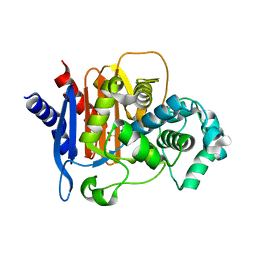 | | Structure of ADC-68, a Novel Carbapenem-Hydrolyzing Class C Extended-Spectrum -Lactamase from Acinetobacter baumannii | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-lactamase ADC-68, CITRIC ACID | | Authors: | Hong, M.K, Kang, L.W. | | Deposit date: | 2014-05-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of ADC-68, a novel carbapenem-hydrolyzing class C extended-spectrum beta-lactamase isolated from Acinetobacter baumannii
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3UU0
 
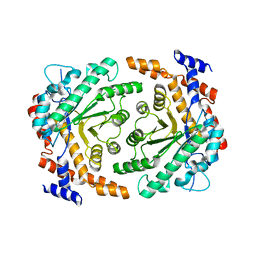 | | Crystal structure of L-rhamnose isomerase from Bacillus halodurans in complex with Mn | | Descriptor: | L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Doan, T.T.N, Prabhu, P, Kim, J.K, Jeya, M, Kang, L.W, Lee, J.K. | | Deposit date: | 2011-11-27 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based studies on the metal binding of two-metal-dependent sugar isomerases.
Febs J., 281, 2014
|
|
3UVA
 
 | | Crystal structure of L-rhamnose isomerase mutant W38F from Bacillus halodurans in complex with Mn | | Descriptor: | L-Rhamnose isomerase, MANGANESE (II) ION | | Authors: | Doan, T.T.N, Prabhu, P, Jeya, M, Kim, J.K, Kang, L.W, Lee, J.K. | | Deposit date: | 2011-11-29 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure-based studies on the metal binding of two-metal-dependent sugar isomerases.
Febs J., 281, 2014
|
|
3UXI
 
 | | Crystal structure of L-rhamnose isomerase W38A mutant from Bacillus halodurans | | Descriptor: | L-Rhamnose isomerase, MANGANESE (II) ION | | Authors: | Doan, T.T.N, Prabhu, P, Kim, J.K, Jeya, M, Kang, L.W, Lee, J.K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-12-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure-based studies on the metal binding of two-metal-dependent sugar isomerases.
Febs J., 281, 2014
|
|
5XD0
 
 | | Apo Structure of Beta-1,3-1,4-glucanase from Paenibacillus sp.X4 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glucanase, TRIETHYLENE GLYCOL | | Authors: | Baek, S.C, Ho, T.-H, Kang, L.-W, Kim, H. | | Deposit date: | 2017-03-24 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Improvement of enzyme activity of beta-1,3-1,4-glucanase from Paenibacillus sp. X4 by error-prone PCR and structural insights of mutated residues.
Appl. Microbiol. Biotechnol., 101, 2017
|
|
4REJ
 
 | |
4REI
 
 | |
1Z32
 
 | | Structure-function relationships in human salivary alpha-amylase: Role of aromatic residues | | Descriptor: | 4-amino-4,6-dideoxy-alpha-D-glucopyranose, 5-HYDROXYMETHYL-CHONDURITOL, CALCIUM ION, ... | | Authors: | Ramasubbu, N, Ragunath, C, Sundar, K, Mishra, P.J, Gyemant, G, Kandra, L. | | Deposit date: | 2005-03-10 | | Release date: | 2005-05-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-function relationships in human salivary alpha-amylase: Role of aromatic residues
To be Published
|
|
4Z5X
 
 | | Glycogen phosphorylase in complex with gallic acid | | Descriptor: | 3,4,5-trihydroxybenzoic acid, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Kantsadi, L.A, Stravodimos, A.G, Chatzileontiadou, S.M.D, Leonidas, D.D. | | Deposit date: | 2015-04-03 | | Release date: | 2015-05-13 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Natural flavonoids as antidiabetic agents. The binding of gallic and ellagic acids to glycogen phosphorylase b.
Febs Lett., 589, 2015
|
|
4YUA
 
 | | Glycogen phosphorylase in complex with ellagic acid | | Descriptor: | 2,3,7,8-tetrahydroxychromeno[5,4,3-cde]chromene-5,10-dione, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Kantsadi, L.A, Stravodimos, A.G, Chatzileontiadou, S.M.D, Leonidas, D.D. | | Deposit date: | 2015-03-18 | | Release date: | 2015-05-13 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Natural flavonoids as antidiabetic agents. The binding of gallic and ellagic acids to glycogen phosphorylase b.
Febs Lett., 589, 2015
|
|
1GY2
 
 | | Crystal structure of Met148Leu rusticyanin | | Descriptor: | COPPER (II) ION, GLYCEROL, RUSTICYANIN | | Authors: | Hough, M.A, Kanbi, L.D, Antonyuk, S, Dodd, F, Hasnain, S. | | Deposit date: | 2002-04-16 | | Release date: | 2002-05-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structures of the met148Leu and Ser86Asp Mutants of Rusticyanin from Thiobacillus Ferrooxidans: Insights Into the Structural Relationship with the Cupredoxins and the Multi Copper Proteins.
J.Mol.Biol., 320, 2002
|
|
1GY1
 
 | | Crystal structures of Ser86Asp and Met148Leu Rusticyanin | | Descriptor: | COPPER (II) ION, RUSTICYANIN | | Authors: | Hough, M.A, Kanbi, L.D, Antonyuk, S, Dodd, F, Hasnain, S. | | Deposit date: | 2002-04-16 | | Release date: | 2002-05-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structures of the met148Leu and Ser86Asp Mutants of Rusticyanin from Thiobacillus Ferrooxidans: Insights Into the Structural Relationship with the Cupredoxins and the Multi Copper Proteins
J.Mol.Biol., 320, 2002
|
|
7KNG
 
 | | 2.10A resolution structure of independent Phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (Ce-2 Y7F) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, DTY-ASP-TYR-PRO-GLY-ASP-PHE-CYS-TYR-LEU-TYR-GLY-THR-CYS, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Weidmann, M, Dranchak, P, Aitha, M, Queme, B, Collmus, C.D, Kanter, L, Lamy, L, Tao, D, Rai, G, Suga, H, Inglese, J. | | Deposit date: | 2020-11-04 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-activity relationship of ipglycermide binding to phosphoglycerate mutases.
J.Biol.Chem., 296, 2021
|
|
7KNF
 
 | | 1.80A resolution structure of independent Phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (Ce-1 NHOH) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, DTY-ASP-TYR-PRO-GLY-ASP-HIS-CYS-TYR-LEU-TYR-GLY-THR, SODIUM ION, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Weidmann, M, Dranchak, P, Aitha, M, Queme, B, Collmus, C.D, Kanter, L, Lamy, L, Tao, D, Rai, G, Suga, H, Inglese, J. | | Deposit date: | 2020-11-04 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-activity relationship of ipglycermide binding to phosphoglycerate mutases.
J.Biol.Chem., 296, 2021
|
|
